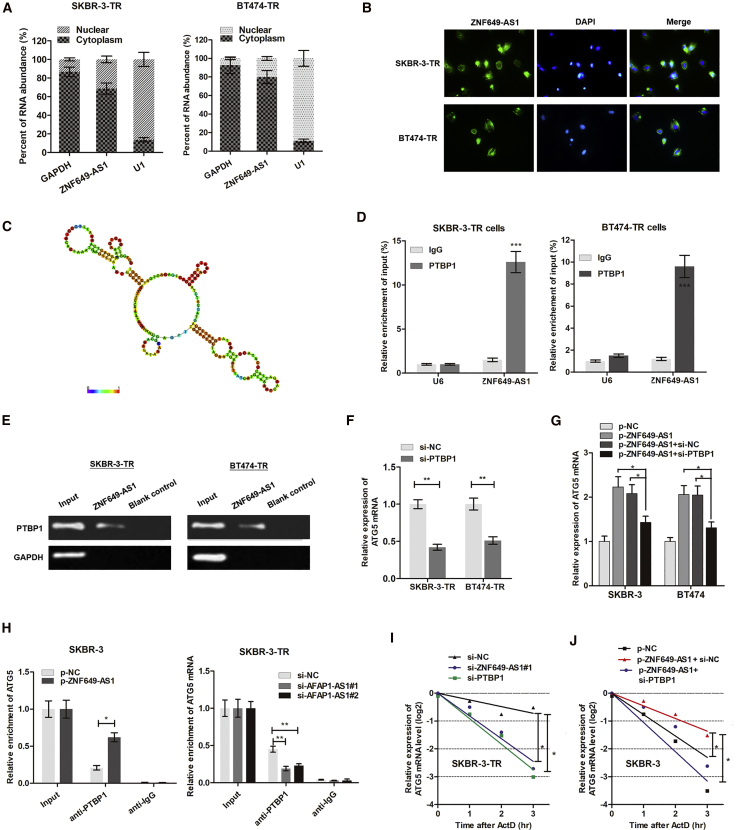
Figure 7.
ZNF649-AS1 Activates the Transcription of ATG5 via Recruiting PTBP1
(A) Cellular fractionation PCR showed that ZNF649-AS1 was mainly distributed in the cytoplasm section. (B) RNA fluorescence in situ hybridization (RNA-FISH) verified that ZNF649-AS1 was mainly distributed in cytoplasm of SKBR-3-TR and BT474-TR cells. (C) Prediction of 981–1190 nt ZNF649-AS1 structure was based on minimum free energy (MFE) and partition function (http://rna.tbi.univie.ac.at/). (D) RNA pull-down assay showed that PTBP1 protein was directly enriched by ZNF649-AS1 probe in breast cancer cells. (E) RIP was performed using anti-PTBP1 and control IgG antibodies, followed by quantitative real-time PCR to examine the enrichment of ZNF649-AS1 and U6. U6 served as a negative control. (F) ATG5 mRNA was silenced by knockdown of PTBP1. (G) Quantitative real-time PCR showed that ZNF649-AS1 upregulated ATG5 expression; however, this upregulation was significantly reversed by co-transfection of si-PTBP1. (H) The endogenous PTBP1 binding to ATG5 mRNA was increased by overexpression of ZNF649-AS1 in SKBR-3 cells, while it was decreased by knockdown of ZNF649-AS1 in SKBR-3-TR cells. (I and J) SKBR-3-TR (I) and SKBR-3 (J) cells silenced or overexpressed with ZNF649-AS1 were cultured with 2 μg/mL actinomycin D (ActD) for 1–3 h, then subjected to qPCR analysis of ATG5. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.