Abstract
Macrophages are professional phagocytic cells that play a critical role in initiating immune responses by presenting antigen and phagocytic clearance. The macrophages can be targeted for immunomodulation by beneficial microbes, such as probiotics. The aim of this study is to investigate the protective effect of Saccharomyces boulardii against Clostridium perfringens infection in avian macrophage cell line HD11. In this study, HD11 macrophages were prestimulated with S. boulardii for 6 h and then infected with C. perfringens for 3 h. Results showed that S. boulardii enhanced phagocytosis and bactericidal capacity against C. perfringens by HD11 cells. The S. boulardii effectively promoted the mRNA expression of CD80, CD83, and CD197 cell-surface molecules in C. perfringens-infected HD11 cells. Moreover, we found that prestimulation with S. boulardii reduced the mRNA expression of CD40, toll-like receptor [TLR] 4, and TLR15 induced by C. perfringens and thereby downregulated the mRNA expression of myeloid differentiation primary response 88, TNF receptor associated factor 6, nuclear factor kappa-B p65 subunit, and c-Jun N-terminal kinase genes in HD11 cells. The upregulation of cytokines (interleukin [IL]-6, tumor necrosis factor alpha, and IL-10) and inducible nitric oxide synthase mRNA expression in C. perfringens-infected HD11 cells were noticeably inhibited by S. boulardii pretreatment. Conclusively, these results might provide a new insight into the role of S. boulardii in regulating avian immune defense against C. perfringens invasion and immune escape.
Key words: Saccharomyces boulardii, HD11 macrophage, immunomodulation, Clostridium perfringens
Introduction
Clostridium perfringens (C. perfringens) is an anaerobic spore-forming pathogenic bacterium that causes a wide variety of diseases ranging from gas gangrene and food poisoning in humans to necrotic enteritis in animals (Kiu and Hall, 2018). It is commonly found in soil, dust, drinking water, feed, litter, feces, wastewater, and the gastrointestinal tract of humans and animals as a normal component of microbiota (Fasina et al., 2016; Prescott et al., 2016). Normally, C. perfringens does not cause intestinal pathophysiological changes and gut microbiota imbalance of broilers (Lacey et al., 2016; Schoster et al., 2019). However, predisposing factors, such as intestinal mucosal damage induced by Eimeria spp. and virus, high dietary levels of animal protein (e.g., fish meal) and fat, intestinal microbiota imbalance, or improper management (e.g., overcrowding), may favor proliferation of C. perfringens and secretion of its toxins that induce intestinal mucosal damage in chickens, and thereby results in subclinical or clinical necrotic enteritis disease (Wu et al., 2014; Moore, 2016). It is reported that the total global economic loss caused by necrotic enteritis is estimated to be over $6 billion per year (Wade et al., 2015). With the prohibitions on the antibiotic growth promoters, proper alternatives such as dietary interventions are being evaluated to improve animal gut health and prevent C. perfringens infection (M'Sadeq et al., 2015). Notable among the feed interventions are the use of direct-fed microbials as feed additives in poultry industry, as they are considered to be “Generally Recognized as Safe” additives and have been shown to be effective pathogen inhibitors and immune modulators in response to pathogenic bacterial infections (Buntyn et al., 2016).
As an antigen-presenting cell, the macrophages play an important role in host defense, tissue repair, and homeostasis (Mao et al., 2015). It is reported that macrophages are plastic, dynamic, and heterogeneous phagocytes (Mills, 2012). On one hand, macrophages phagocytize and kill pathogens by secreting proinflammatory cytokines (e.g., interleukin [IL]-6, IL-12, tumor necrosis factor alpha [TNF-α], and IL-1β), chemokines (e.g., CCL2, CCL5, and CXCL8), and chemical species (reactive oxygen species, reactive nitrogen species) to induce inflammation (Benoit et al., 2008; Murray, 2017). On the other hand, macrophages secrete anti-inflammatory cytokines (e.g., IL-4, IL-10, and TGF-β) and growth factors (e.g., PDGF, VEGF, and EGF) to dampen excessive or prolonged inflammation which can result in tissue injury and contribute to pathogenesis (Vannella and Wynn, 2017). Depending on the converging signals from inflammatory stimuli and the cellular environment, macrophages activation is broadly described as M1 and M2 phenotype (Vergadi et al., 2017). M1 phenotypic macrophages can be activated by bacteria-derived lipopolysaccharide (LPS) via toll-like receptors/myeloid differentiation primary response 88 (TLR/MyD88) signaling pathway, which has shown to be of vital importance in controlling intracellular pathogens (Da Silva et al., 2007; Mosser and Edwards, 2008).
Probiotics are live microorganisms, which exert a beneficial influence on animal health by reshaping gut microbiota, enhancing digestive capacity, improving gastrointestinal mucosal barrier, inhibiting intestinal pathogenic bacteria growth and colonization, or regulating mucosal immune system (Sánchez et al., 2017). Several selected probiotics, such as lactic acid bacteria, Bacillus species, Bifidobacterium, and yeast, have been applied to improve animal performance and health in food animal production (Wang et al., 2016). A number of studies in vivo have shown that Saccharomyces boulardii, as a nonpathogenic yeast, has been introduced to prevent or treat various gastrointestinal complications both in human and animals (Anand et al., 2018). Our previous in vivo study showed that S. boulardii supplementation modulates intestinal ultrastructure by upregulating the tight junction protein expression in broilers (Rajput et al., 2013). However, the role of S. boulardii on C. perfringens-induced inflammatory response in chicken macrophages is less studied, and the mechanism remains obscure. In this study, we investigated the protective effect of S. boulardii against C. perfringens infection in avian HD11 cell lines and its underlying immune mechanism.
Materials and methods
Strains and Culture Conditions
S. boulardii was isolated and identified by our lab and was cultured overnight at 30°C in yeast peptone dextrose broth (OXOID; UK) under aerobic conditions. Clostridium perfringens type A (ATCC 13124) was cultured in reinforced clostridium medium at 37°C for 20 h in anaerobic gas generating packs (Mitsubishi Gas Chemical Company Inc., Tokyo, Japan). S. boulardii and C. perfringens were harvested by centrifugation at 3,500 × g for 10 min, respectively. After 3 times washing with phosphate-buffered saline (PBS, pH = 7.2), the pellets of S. boulardii and C. perfringens were resuspended in RPMI 1640 medium (antibiotics free) for preincubation or infection. Optical density method (SpectraMax M5, Molecular Devices, San Jose, CA) was performed to adjust the final concentration of S. boulardii and C. perfringens.
Cell Culture
The avian macrophage cell line HD11 was kindly provided by Dr. Shou-Qun Jiang (GDAAS, Guangzhou, China) and was cultured in RPMI 1640 medium (Gibco, Carlsbad, CA) supplemented with 10% chicken serum (Gibco), 100 μg/mL streptomycin, and 100 U/mL penicillin (Sigma-Aldrich, St. Louis, MO), 1× nonessential amino acids (Invitrogen, Waltham, MA), sodium pyruvate (1 mmol/L), L-glutamine (2 mmol/L), and 2-mercaptoethanol (5 × 10−5 mol) at 41°C in a 5% humidified CO2 incubator. If not mentioned, the antibiotics (100 μg/mL streptomycin and 100 U/mL penicillin) will not add into the RPMI 1640 medium in the further experiment.
Cytotoxicity Assay
Cell viability was determined by cell counting kit-8 (CCK-8, Beyotime, Nanjing, China) according to the manufacturer's instruction. Briefly, HD11 cells were seeded into 96-well microplate at 2.0 × 10ˆ4 cells/mL (Corning Inc., Corning, NY) and incubated with S. boulardii (multiplicity of infection (MOI) = 0, 100, 1,000, 5,000, 10,000) for 6 h. After washed 3 times with sterile PBS, the cell wells were added with CCK-8 kit solution (10 μL/well) and further incubated for 1 h. Subsequently, the optical density value was measured by SpectraMax M5 (Molecular Devices) at OD450, and the relative cell viabilities was calculated as previously described (Mosmann, 1983). The release of lactate dehydrogenase (LDH) from the damaged cells was quantified using LDH kit (Beyotime) after HD11 cells incubated with S. boulardii (MOI = 0, 1, 10, 100) for 6 h (Wu et al., 2017).
Clostridium perfringens-Killing Analysis
The effect of S. boulardii on the C. perfringens-killing capacity of chicken macrophage HD11 cells was measured by a viable count method, as described previously (Wang et al., 2018). Briefly, HD11 cells seeded into 12-well plates (2 × 10ˆ5 cells/mL) were preincubated with S. boulardii (MOI = 100, 2 × 10ˆ7 CFU/well) for 6 h under aerobic conditions. For phagocytosis analysis, after washed 3 times with sterile PBS, HD11 cells were then infected with C. perfringens (MOI = 100, 2 × 10ˆ7 CFU/well) in the RPMI 1640 medium (absence of chicken serum) at 41°C for 1 h under aerobic conditions. For bactericidal analysis, the infected HD11 cells were washed 3 times with sterile PBS and then incubated in RPMI 1640 containing gentamicin (25 μg/mL) and ampicillin (50 μg/mL) for another 24 h. At each of time points (0 h and 24 h), infected HD11 cells were washed 5 times with sterile PBS and lysed with 0.01% Triton X-100 diluted in PBS (1 mL/well). The serial 10-fold dilutions of cell lysates were immediately plated on reinforced clostridium medium agar plates in triplicate and then incubated at 37°C for 24 h under anaerobic conditions. The number of surviving intracellular bacteria was determined by bacterial colony counting in the next day.
RNA Isolation and Quantitative Real-Time PCR
HD11 cells seeded into 12-well plates (2 × 10ˆ5 cells/mL) were pretreated with S. boulardii (MOI = 100) for 6 h, and then infected with C. perfringens (MOI = 100) for 3 h, and then washed by PBS 3 times to collect the cell pellets. Total RNA isolated from HD11 cells using RNAiso Plus (TaKaRa, Dalian, China) was reverse-transcribed by PrimeScript II first Strand cDNA Synthesis Kit (TaKaRa). Reverse transcription-qPCR was performed using SYBR PremixExTaq II (TaKaRa) and the StepOne Plus Real-Time PCR system (Applied Biosystems, Carlsbad, CA). All primer sequences for chicken macrophages genes are listed in Table 1. The housekeeping gene β-actin was selected as reference gene, and relative quantification was calculated using the 2−ΔΔCq method (Bustin et al., 2009). ΔCq is Cq, target − Cq, reference, and ΔΔCq is ΔCq, treatment − ΔCq, control.
Table 1.
Primer sequences used for qRT-PCR.
| Gene name | Primer sequence (5′-3′) | Product size | Accession No. |
|---|---|---|---|
| CD40 | F: GGCACCTTCTCCAATGTATCTTC | 96 | NM_204665 |
| R: GTTCGTCCCTTTCACCTTCAC | |||
| CD80 | F: CAGCAAGCCGAACATAGAAAGA | 270 | NM_001079739 |
| R: AGCAAACTGGTGGACCTGAGA | |||
| CD83 | F: GCTGACTTGCCTCGGGATT | 272 | XM_418929.5 |
| R: TCACTCCGCTATCCGTCTCA | |||
| CD197 | F: GACGACTATGACGCCAACAC | 211 | NM_001198752 |
| R: CCAGGTTCAGCAAGTAGATGTC | |||
| IL-6 | F: CTCCTCGCCAATCTGAAGTC | 99 | NM_204628 |
| R: CCTCACGGTCTTCTCCATAAAC | |||
| IL-1β | F: CGACATCAACCAGAAGTGCTT | 298 | NM_204524 |
| R: GTCCAGGCGGTAGAAGATGA | |||
| TNF-α | F: GGACAGCCTATGCCAACAAG | 81 | NM_204267 |
| R: GCGGTCATAGAACAGCACTAC | |||
| IL-10 | F: ACCAGTCATCAGCAGAGCAT | 222 | NM_001004414 |
| R: CCTCCTCATCAGCAGGTACTC | |||
| iNOS | F: TACTCTTGGCGTCATTACTC | 67 | NM_204961 |
| R: GCATAGATCACAGTCACCTT | |||
| TLR1 | F: CCGTTCAAGTGTTCGTGTGA | 116 | NM_001007488 |
| R: CCGCTCAAGTCTTCTGGGTA | |||
| TLR2 | F: TGTTCCTGTTCATCCTCATCCT | 168 | NM_204278 |
| R: AGTTGGAGTCGTTCTCACTGT | |||
| TLR4 | F: GAATGACACGGACACTCTT | 95 | NM_001030693 |
| R: ACATAGGAACCTCTGACAAC | |||
| TLR15 | F: CTTGTCGTTCTGGTGCTAA | 156 | NM_001037835 |
| R: ATCGTGCTCGCTGTATGA | |||
| MyD88 | F: GGATGGTGGTCGTCATTTCA | 225 | NM_001030962 |
| R: GAGATTTTGCCAGTCTTGTCCA | |||
| TRAF6 | F: ATATCCAGTTACCAAGTGCTCAG | 91 | XM_004941548 |
| R: CAAGGCAGATGGCTGTCATAT | |||
| NF-κB p65 | F: CTTCAATGTGCCAATGGAGGAG | 271 | NM_205129 |
| R: CTCAGCCCAGAAACGAACCT | |||
| JNK | F: GTAGTAGATCCAGACAAGAGAA | 71 | NM_205095 |
| R: TCATACCAGACCGTAATATAGG | |||
| β-actin | F: TATGTGCAAGGCCGGTTTC | 110 | NM_205518 |
| R: TGTCTTTCTGGCCCATACCAA |
Abbreviations: CD, cluster of differentiation; F, forward; IL, interleukin; iNOS, inducible nitric oxide synthase; JNK, c-Jun N-terminal kinase; MyD88, myeloid differentiation primary response 88; NF-κB p65, nuclear factor kappa-B p65 subunit; R, reverse; TLR, toll like receptor; TNF-α, tumor necrosis factor alpha; TRAF6, TNF receptor associated factor 6.
Statistical Analysis
Results between 2 groups were analyzed by 2-tailed Student t test using SPSS 22.0 (SPSS Inc., Chicago, IL), and graphs were represented as mean ± SD of at least 3 independent experiments and visualized by Origin 8.0 (OriginLab, Berkeley, CA). ∗P < 0.05, ∗∗P < 0.01.
Results
Cytotoxicity Analysis of S. boulardii
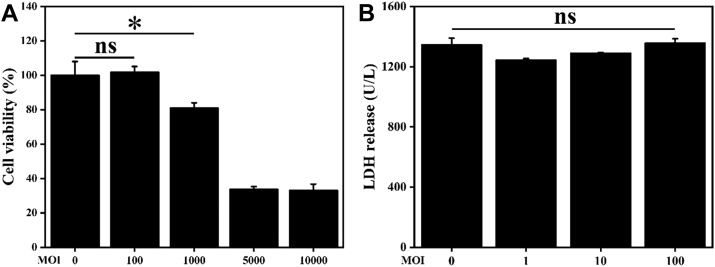
The cytotoxicity of S. boulardii on chicken HD11 cells was measured by the CCK-8 kit. Significant (P < 0.05) cell cytotoxicity was observed when macrophages were incubated with S. boulardii at MOI of 1,000 but not at MOI of 100 (P > 0.05, Figure 1A). Moreover, the safety of S. boulardii at MOI of 100 was further confirmed by testing the release of the cytosolic marker LDH (Korzeniewski and Callewaert, 1983). There was no significant (P > 0.05) difference in LDH release among the groups (Figure 1B). Therefore, S. boulardii at MOI of 100 was selected for the further experiments.
Figure 1.
Cytotoxicity analysis of S. boulardii on chicken HD11 cells. HD11 cells were incubated with PBS or S. boulardii (MOI = 0, 100, 1,000, 5,000 or 10,000) for 6 h. (A) Cell viability was determined by the CCK-8 assay. (B) Cell damage was measured by testing the release of LDH. Results are presented as mean ± SD of 8 (CCK-8 assay) or 6 (LDH assay) samples. ∗P < 0.05 (t test). ns indicates no significance (P > 0.05). Abbreviations: LDH, lactate dehydrogenase; MOI, multiplicity of infection.
S. boulardii Enhances Phagocytosis and Intracellular Bactericidal Capacity of Macrophages
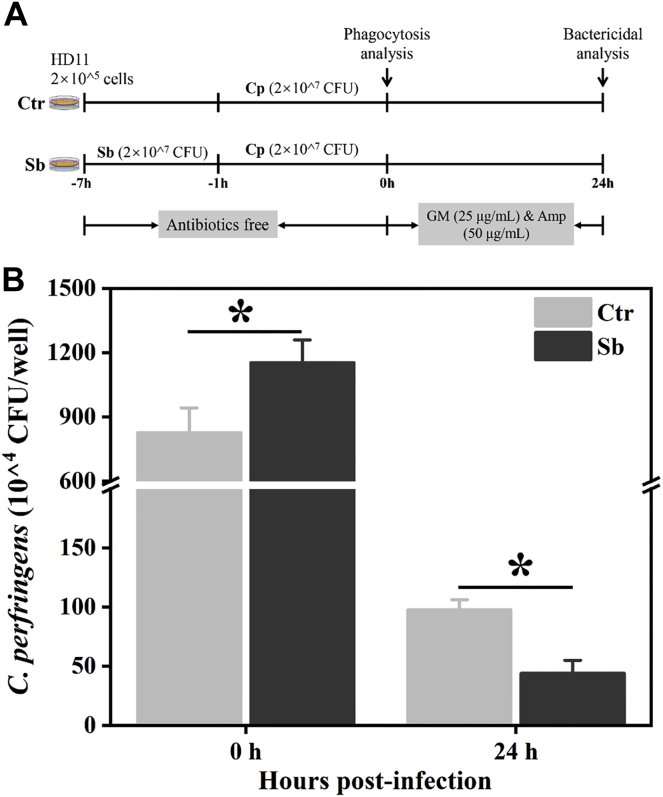
Phagocytosis and intracellular bactericidal capacity play vital roles in macrophage-mediated host defense, which leads to internalization and death of pathogens (Mao et al., 2015). We further examined whether S. boulardii improved the phagocytosis and intracellular bactericidal capacity of the HD11 macrophages. The results showed that S. boulardii treatment significantly (P < 0.05) increased the uptake of C. perfringens in HD11 cells (Figure 2B, 0 h), suggesting that S. boulardii enhances the phagocytosis of HD11 cells. After 24 h incubation, the intracellular survival of C. perfringens in S. boulardii-pretreated HD11 cells significantly (P < 0.05) reduced compared with the unpretreated macrophages (Figure 2B, 24 h). The results demonstrated that S. boulardii could enhance phagocytosis and intracellular bactericidal capacity of avian macrophages.
Figure 2.
S. boulardii enhances phagocytosis and bactericidal capacity of HD11 cells. (A) Schematic illustration of C. perfringens-killing assay. HD11 cells were pretreated with S. boulardii (MOI = 100) for 6 h. After being infected with C. perfringens (MOI = 100) for 1 h, washed, and incubated in RPMI 1640 medium with gentamicin (25 μg/mL) and ampicillin (100 μg/mL) for 0 h or 24 h, these cells were lysed, diluted, and plated on reinforced clostridium medium (RCM) agar plates for colony enumeration. (B) S. boulardii enhances phagocytosis and bactericidal activity of macrophages. Results are mean ± SD for 3 independent experiments. ∗P < 0.05 (t test). Abbreviations: Amp, ampicillin; Cp, C. perfringens; Ctr, control group; GM, gentamicin; MOI, multiplicity of infection; Sb, S. boulardii treated group.
Effect of S. boulardii on Cell-Surface Molecules mRNA Expression of Macrophages
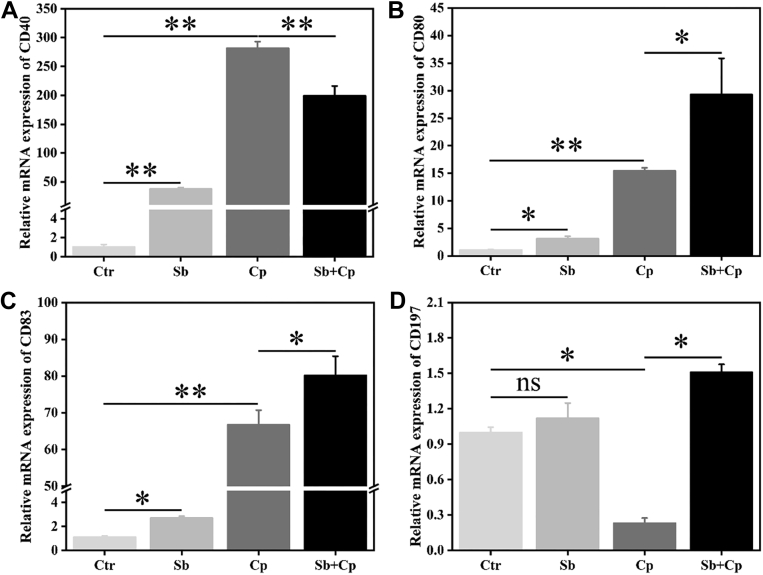
S. boulardii-induced phagocytosis and bactericidal capacity were confirmed by the activation of macrophages. As shown in Figure 3, both S. boulardii and C. perfringens exposure markedly (P < 0.05 or P < 0.01) upregulated the mRNA expression of CD40, CD80, and CD83, whereas C. perfringens significantly (P < 0.05) inhibited the CD197 mRNA expression in HD11 cells. S. boulardii pretreatment significantly (P < 0.05) enhanced C. perfringens-induced mRNA expression of CD80, CD83, and CD197, whereas significantly (P < 0.05) decreased C. perfringens-induced mRNA expression of CD40 in HD11 cells (Figure 3).
Figure 3.
Effect of S. boulardii on cell surface molecule mRNA expression of HD11 macrophages. HD11 cells were preincubated with S. boulardii (MOI = 100) for 6 h and then infected with C. perfringens (MOI = 100) for 3 h. Total RNA was isolated, and the mRNA expression of cluster of differentiation 40 (CD40) (A), CD80 (B), CD83 (C), and CD197 (D) was analyzed by real-time PCR. Results are mean ± SD for 3 independent experiments. ∗P < 0.05, ∗∗P < 0.01 (t test). ns indicates no significance (P > 0.05). Abbreviations: Cp, C. perfringens treated group; Ctr, control group; MOI, multiplicity of infection; Sb, S. boulardii treated group; Sb + Cp, S. boulardii pretreated and then Cp infected group.
Effect of S. boulardii on mRNA Expression of TLR Signaling Molecules in Macrophages
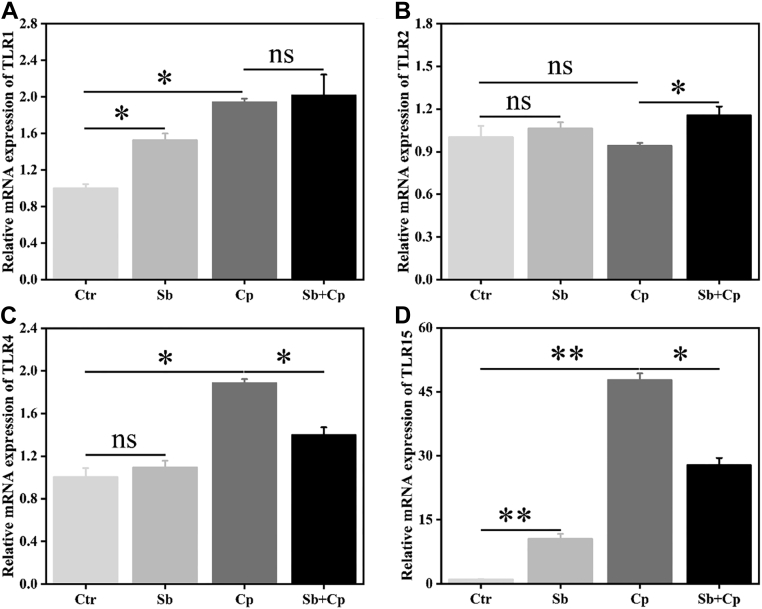
The TLR signaling pathways play an important role in the initiation and activation of macrophage inflammatory responses (Fitzgerald and Kagan, 2020). As shown in Figure 4, both S. boulardii and C. perfringens exposure significantly enhanced TLR1 and TLR15 mRNA expression (P < 0.05 and P < 0.01, respectively), and C. perfringens infection upregulated (P < 0.05) the TLR4 mRNA expression in HD11 cells. S. boulardii pretreatment markedly (P < 0.05) downregulated the mRNA expression of TLR4 and TLR15 induced by C. perfringens, whereas upregulated (P < 0.05) TLR2 mRNA expression in HD11 cells.
Figure 4.
Effects of S. boulardii on TLRs mRNA expression of HD11 macrophages. HD11 cells were preincubated with S. boulardii (MOI = 100) for 6 h and then infected with C. perfringens (MOI = 100) for 3 h. Then, total RNA was isolated, and the mRNA expression of toll like receptor 1 (TLR1) (A), TLR2 (B), TLR4 (C), and TLR15 (D) was analyzed by real-time PCR. Results are mean ± SD for 3 independent experiments. ∗P < 0.05, ∗∗P < 0.01 (t test). ns indicates no significance (P > 0.05). Abbreviations: Cp, C. perfringens treated group; Ctr, control group; MOI, multiplicity of infection; Sb, S. boulardii treated group; Sb + Cp, S. boulardii pretreated and then Cp infected group.
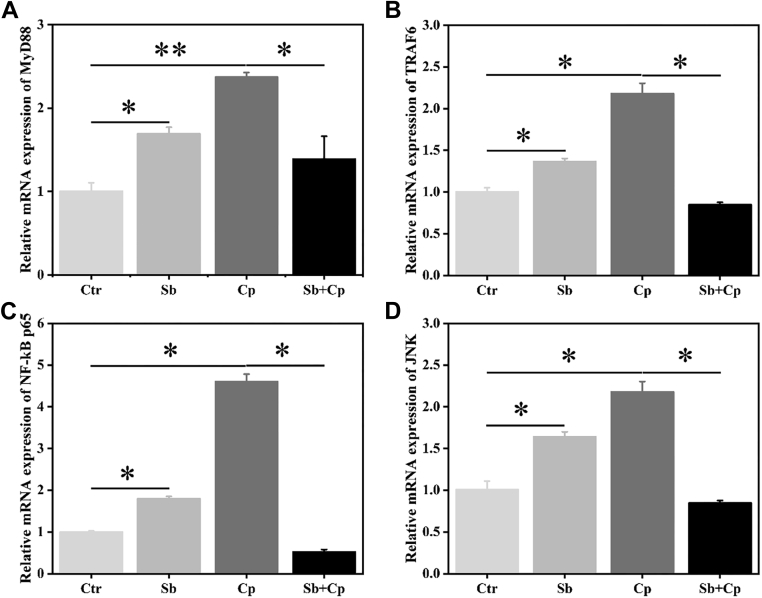
As shown in Figure 5, both S. boulardii and C. perfringens exposure significantly (P < 0.05 or P < 0.01) enhanced the mRNA expression of MyD88, TNF receptor associated factor 6 (TRAF6), nuclear factor kappa-B p65 subunit (NF-κB p65), and c-Jun N-terminal kinase (JNK), whereas S. boulardii pretreatment markedly (P < 0.05) downregulated C. perfringens-induced the mRNA expression of MyD88, TRAF6, NF-κB p65, and JNK in macrophages (Figure 5). To sum up, these data demonstrated that S. boulardii pretreatment inhibited C. perfringens-induced activation of TLR4/TLR15-MyD88 signaling pathways and thus inflammation.
Figure 5.
Effect of S. boulardii on the signaling pathways related mRNA expression of HD11 macrophages. HD11 cells were preincubated with S. boulardii (MOI = 100) for 6 h and then infected with C. perfringens (MOI = 100) for 3 h. Then total RNA was isolated and the mRNA expression of MyD88 (A), TRAF6 (B), NF-κB p65 (C), and JNK (D) was analyzed by real-time PCR. Results are mean ± SD for 3 independent experiments. ∗P < 0.05, ∗∗P < 0.01 (t test). Abbreviations: Cp, C. perfringens treated group; Ctr, control group; JNK, c-Jun N-terminal kinase; MOI, multiplicity of infection; MyD88, myeloid differentiation primary response 88; NF-κB p65, nuclear factor kappa-B p65 subunit; Sb, S. boulardii treated group; Sb + Cp, S. boulardii pretreated and then Cp infected group; TRAF6, TNF receptor associated factor 6.
Effect of S. boulardii on mRNA Expression of Inflammatory Factors in Macrophages
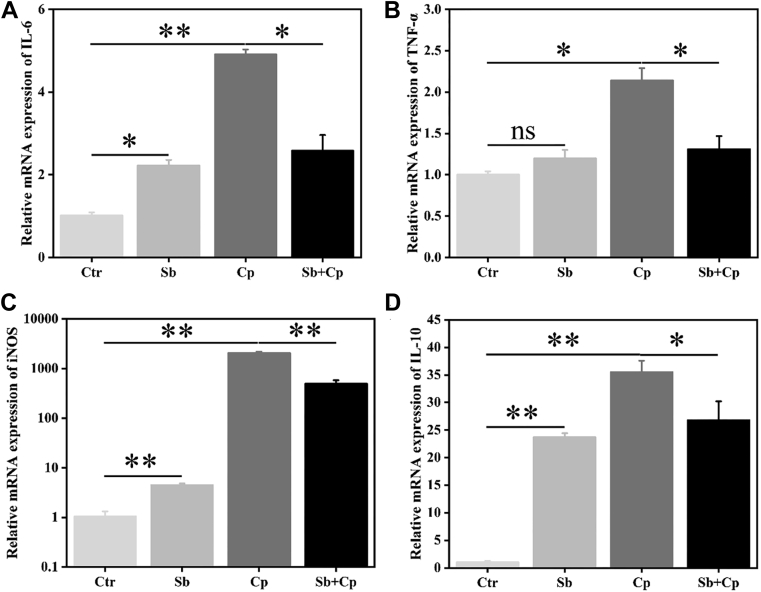
The downstream target mRNA expression of TLRs signaling pathways was further measured by qPCR (Figure 6). The results showed that both S. boulardii and C. perfringens exposure significantly (P < 0.05 or P < 0.01) upregulated the mRNA expression of IL-6, inducible nitric oxide synthase (iNOS), and IL-10 in HD11 cells, whereas C. perfringens also significantly increased the mRNA expression of TNF-α (P < 0.05). Additionally, S. boulardii pretreatment significantly (P < 0.05) reduced C. perfringens-induced mRNA expression of IL-6, TNF-α, iNOS, and IL-10 in HD11 cells. These results indicated that S. boulardii alleviates C. perfringens-induced excessive proinflammatory response.
Figure 6.
Effect of S. boulardii on inflammatory factors mRNA expression of HD11 macrophages. HD11 cells were preincubated with S. boulardii (MOI = 100) for 6 h and then infected with C. perfringens (MOI = 100) for 3 h. Then total RNA was isolated, and the mRNA expression of IL-6 (A), TNF-α (B), iNOS (C), and IL-10 (D) was analyzed by real-time PCR. Results are mean ± SD for 3 independent experiments. ∗P < 0.05, ∗∗P < 0.01 (t test). ns indicates no significance (P > 0.05). Abbreviations: Cp, C. perfringens treated group; Ctr, control group; IL, interleukin; iNOS, inducible nitric oxide synthase; MOI, multiplicity of infection; Sb, S. boulardii treated group; Sb + Cp, S. boulardii pretreated and then Cp infected group; TNF-α, tumor necrosis factor alpha.
Discussion
Macrophages play indispensable roles in initiating the innate immune response, shaping the adaptive immune response, and the defense against pathogens (Hoebe et al., 2004). Once stimulated by foreign antigens and inflammatory stimuli, immature macrophages undergo maturation, evolving from antigen-presenting cells to T cell-priming cells to promote T lymphocyte activation (Laskin, 2009). Previous studies in vivo have evidenced that probiotics, including S. boulardii, have a beneficial effect in improving growth performance and prevention of pathogenic infections in poultry (Li et al., 2018; de Oliveira et al., 2019; Nari and Ghasemi, 2020; Ramlucken et al., 2020). However, the protective mechanism of S. boulardii mediated by chicken macrophages remains unclear. Phagocytosis by macrophages is critical for the uptake and degradation of intracellular pathogens and for the initiation of the innate immune response (Aderem and Underhill, 1999). However, some pathogens, such as Listeria, Salmonella typhimurium and Clostridium perfringens, can survive and replicate in macrophages by escaping the phagosome and modifying the vacuolar maturation process of macrophages (Aderem and Underhill, 1999; O'Brien and Melville, 2004). Previous studies found that Bacillus amyloliquefaciens could enhance the phagocytosis and bactericidal activity of murine macrophages against S. Typhimurium or E.coli (Wu et al., 2017; Fu et al., 2019), which was consistent with our results. The present results showed that S. boulardii enhanced the HD11 macrophages' phagocytosis and its ability to clear intracellular C. perfringens (Figure 2), which was further confirmed by the activation and maturation of the macrophages (Figure 3). Activated macrophages can be characterized by the high expression of costimulatory molecules (CD40, CD80, and CD83) or surface marker (CD197), which are necessary for the activation of T and B lymphocytes in the host (Zotos and Tarlinton, 2012; Wang et al., 2018). The present results showed that S. boulardii pretreatment promoted the C. perfringens-induced activation and maturation of HD11 macrophages, as evidenced by the upregulation of macrophage surface marker (CD197) and costimulatory receptors (CD80 and CD83) mRNA expression, consistent with previous observations (Wang et al., 2018). These results indicate that S. boulardii activates an innate immune response in HD11 macrophages to eliminate intracellular C. perfringens. CD40 (also known as tumor necrosis factor receptor superfamily 5) is a member of the tumor necrosis factor receptor superfamily that is expressed on macrophages, dendritic cells, and B cells (Croft et al., 2013). Not only is CD40 essential for activation and proliferation of B cells by T cell-dependent antigens, but also it is an important regulator to induce the production of inflammatory cytokines in macrophages and dendritic cells (Quezada et al., 2004; Croft et al., 2013; Croft and Siegel, 2017). In the present study, S. boulardii pretreatment downregulated C. perfringens-induced CD40 mRNA expression (Figure 3A), indicating that S. boulardii might attenuate C. perfringens-induced inflammatory response by blocking CD40.
Inflammatory response is regulated by a complex and cross-linked endogenous cellular signaling pathways and their modulators (Takeuchi and Akira, 2010). The immune responses of the host are initiated immediately after pattern recognition receptors recognize pathogen-associated molecular patterns, which is essential for effective clearance of infectious microbes (Palm and Medzhitov, 2009). Evidence has shown that the host–microbe interactions depend on the pattern recognition receptors, such as the TLRs and the pathogen-associated molecular patterns (Fitzgerald and Kagan, 2020). TLR2 can recognize many different microbial and synthetic components (Beutler et al., 2006), whereas TLR1/2 heterodimers recognize di- or tri-acylated lipoproteins or lipopeptides (Jin et al., 2007). Although TLR4 specifically recognizes bacteria-derived LPS (Lee and Kim, 2007), many studied have recorded that TLR4 is also involved in the host immune responses induced by C. perfringens. Shi et al. (2019) reported that TLR4/MyD88/NF-κB signaling pathway was involved in the inflammatory responses in piglet diarrhea induced by C. perfringens. C. perfringens α-toxin disturbs host defense by modulating TLR4-mediated inflammatory response (Takehara et al., 2019). Avian-specific TLR15 recognizes nonsecreted, heat-stabile components of both gram-positive and gram-negative pathogens and plays a constitutive role in the immune defense of chickens (Higgs et al., 2006; Nerren et al., 2010). In the current study, we found that S. boulardii treatment specifically upregulated TLR1 and TLR15 mRNA expression (Figure 4). The constant stimulation of TLR is necessary for maintaining immune homeostasis in the intestine (Rajput et al., 2014). We found that C. perfringens infection significantly increased the mRNA expression of TLR1, TLR4 and TLR15, whereas S. boulardii pretreatment markedly decreased C. perfringens-induced mRNA expression of TLR4 and TLR15 (Figure 4). After recognition, TLR trigger the activation of the downstream signaling pathways (such as mitogen-activated protein kinase and transcription factors nuclear factor [NF]-κB) by MyD88-dependent and TRIF-dependent (MyD88-independent) ways, which leads to the production of inflammatory cytokines and chemokines (Lee and Kim, 2007). Our data demonstrated that S. boulardii treatment significantly increased the expression of MyD88, TRAF6, NF-κB p65, and JNK (Figure 5), indicating that S. boulardii activated HD11 macrophages via TLR1/TLR15-MyD88-dependent manner. In addition, we also found that S. boulardii pretreatment downregulated C. perfringens-induced overexpression of MyD88, TRAF6, NF-κB p65, and JNK (Figure 5), indicating that S. boulardii attenuated C. perfringens-induced TLR4/TLR15-MyD88 signaling.
The main role of the TLR-MyD88-dependent pathway is to induce the expression of inflammatory cytokines (such as IL-6, IL-12, and TNF-α), which is necessary for pathogen clearance (Lee and Kim, 2007). However, exaggerated immune responses can be detrimental to the host (Kawasaki and Kawai, 2014). Therefore, the excessive immune responses are tightly controlled by associated negative feedback loops and anti-inflammatory cytokines (Martinez et al., 2009; Ip et al., 2017). Evidences have shown that IL-10 and TGF-β are the 2 main potent anti-inflammatory cytokines (Opal and DePalo, 2000). In our study, we found that S. boulardii treatment significantly increased the mRNA expression of both pro- and anti-inflammatory cytokines (IL-6, iNOS, and IL-10) (Figure 6), suggesting that S. boulardii might serve as a regulator to maintain the anti- and pro-inflammatory response in macrophages. Furthermore, we also found that C. perfringens infection significantly upregulated the mRNA expression of inflammatory cytokines (IL-6, TNF-α, iNOS, and IL-10), whereas S. boulardii pretreatment decreased C. perfringens-induced inflammatory cytokines mRNA expression (Figure 6). Similar to our results, Thomas et al. (2009) also found that S. boulardii cultured supernatant reduced secretion of proinflammatory cytokines (IL-6 and TNF-α) in LPS-stimulated dendritic cells. These results indicate that S. boulardii alleviates C. perfringens-induced excessive proinflammatory responses mediated by TLR4/TLR15-MyD88 signaling pathway.
Conclusion
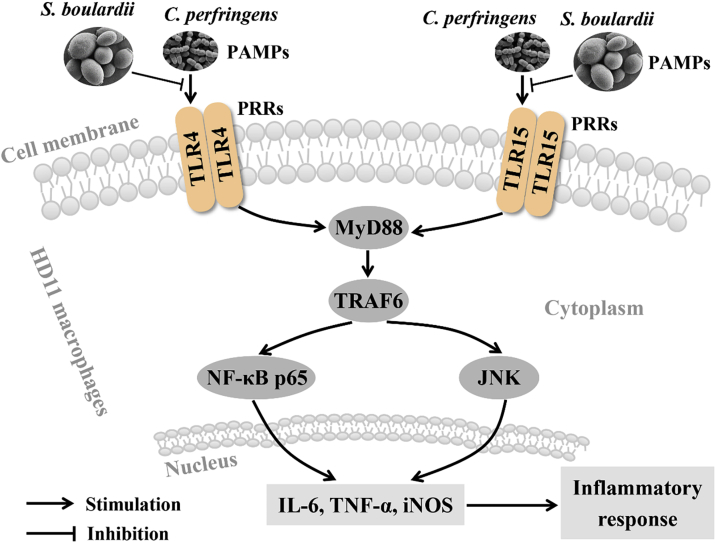
Overall, this study demonstrates that S. boulardii induces HD11 macrophages activation to clear C. perfringens and attenuates C. perfringens-induced inflammatory responses mediated by TLR4/TLR15-MyD88 signaling pathway (Figure 7). These findings might provide a new perspective into the function of S. boulardii in regulating avian immune defense against C. perfringens invasion and immune escape. Given that avian necrotic enteritis is mainly caused by C. perfringens, these findings also provide a potential preventive approach against avian necrotic enteritis caused by C. perfringens. However, further investigations about the protective effect of S. boulardii against C. perfringens infection in broilers are needed.
Figure 7.
Graphical summary of the anti-inflammatory role of Saccharomyces boulardii against Clostridium perfringens infection in chicken HD11 macrophages. S. boulardii induces HD11 macrophages activation to clear C. perfringens and attenuates C. perfringens-induced proinflammatory responses mediated by TLR4/TLR15-MyD88 signaling pathway. Abbreviations: IL, interleukin; iNOS, inducible nitric oxide synthase; JNK, c-Jun N-terminal kinase; MyD88, myeloid differentiation primary response 88; NF-κB p65, nuclear factor kappa-B p65 subunit; PRRs, pattern recognition receptors; TLR, toll like receptor; TNF-α, tumor necrosis factor alpha; TRAF6, TNF receptor associated factor 6.
acknowledgments
This study is jointly supported by the National Natural Science Foundation of China (No. 31472128 and 31672460), the National High-Tech R&D Program Project (863) of China (No. 2013AA102803D), the Key Project of Science and Technology of Zhejiang Province of China (No. 2006C12086), and the Education Foundation of Da Bei Nong Group.
Conflict of Interest Statement: The authors declare that they have no conflict of interest
Contributor Information
Pengwei Zhao, Email: zhaopengwei625@163.com.
Weifen Li, Email: wfli@zju.edu.cn.
References
- Aderem A., Underhill D.M. Mechanisms of phagocytosis in macrophages. Annu. Rev. Immunol. 1999;17:593–623. doi: 10.1146/annurev.immunol.17.1.593. [DOI] [PubMed] [Google Scholar]
- Anand S., Singh K.S., Aggarwal D. Microbial Cell Factories. CRC Press; Boca Raton: 2018. Expanding avenues for probiotic yeast: Saccharomyces boulardii; pp. 125–147. [Google Scholar]
- Benoit M., Desnues B., Mege J.-L. Macrophage polarization in bacterial infections. J. Immunol. 2008;181:3733–3739. doi: 10.4049/jimmunol.181.6.3733. [DOI] [PubMed] [Google Scholar]
- Beutler B., Jiang Z., Georgel P., Crozat K., Croker B., Rutschmann S., Du X., Hoebe K. Genetic analysis of host resistance: toll-like receptor signaling and immunity at large. Annu. Rev. Immunol. 2006;24:353–389. doi: 10.1146/annurev.immunol.24.021605.090552. [DOI] [PubMed] [Google Scholar]
- Buntyn J.O., Schmidt T.B., Nisbet D.J., Callaway T.R. The role of direct-fed microbials in conventional livestock production. Annu. Rev. Anim. Biosci. 2016;4:335–355. doi: 10.1146/annurev-animal-022114-111123. [DOI] [PubMed] [Google Scholar]
- Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L. The MIQE guidelines: minimum information for publication of quantitative real-time pcr experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Croft M., Benedict C.A., Ware C.F. Clinical targeting of the TNF and TNFR superfamilies. Nat. Rev. Drug Discov. 2013;12:147–168. doi: 10.1038/nrd3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft M., Siegel R.M. Beyond TNF: TNF superfamily cytokines as targets for the treatment of rheumatic diseases. Nat. Rev. Rheumatol. 2017;13:217. doi: 10.1038/nrrheum.2017.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Silva F.P., Aloulou M., Skurnik D., Benhamou M., Andremont A., Velasco I.T., Chiamolera M., Verbeek J.S., Launay P., Monteiro R.C. CD16 promotes Escherichia coli sepsis through an FcRγ inhibitory pathway that prevents phagocytosis and facilitates inflammation. Nat. Med. 2007;13:1368–1374. doi: 10.1038/nm1665. [DOI] [PubMed] [Google Scholar]
- de Oliveira M.J.K., Sakomura N.K., de Paula Dorigam J.C., Doranalli K., Soares L., Viana G.D.S. Bacillus amyloliquefaciens CECT 5940 alone or in combination with antibiotic growth promoters improves performance in broilers under enteric pathogen challenge. Poult. Sci. 2019;98:4391–4400. doi: 10.3382/ps/pez223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasina Y.O., Newman M.M., Stough J.M., Liles M.R. Effect of Clostridium perfringens infection and antibiotic administration on microbiota in the small intestine of broiler chickens. Poult. Sci. 2016;95:247–260. doi: 10.3382/ps/pev329. [DOI] [PubMed] [Google Scholar]
- Fitzgerald K.A., Kagan J.C. Toll-like receptors and the control of immunity. Cell. 2020;180:1044–1066. doi: 10.1016/j.cell.2020.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu A., Mo Q., Wu Y., Wang B., Liu R., Tang L., Zeng Z., Zhang X., Li W. Protective effect of Bacillus amyloliquefaciens against Salmonella via polarizing macrophages to M1 phenotype directly and to M2 depended on microbiota. Food Funct. 2019;10:7653–7666. doi: 10.1039/c9fo01651a. [DOI] [PubMed] [Google Scholar]
- Higgs R., Cormican P., Cahalane S., Allan B., Lloyd A.T., Meade K., James T., Lynn D.J., Babiuk L.A., O'Farrelly C. Induction of a novel chicken Toll-like receptor following Salmonella enterica serovar Typhimurium infection. Infect. Immun. 2006;74:1692–1698. doi: 10.1128/IAI.74.3.1692-1698.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoebe K., Janssen E., Beutler B. The interface between innate and adaptive immunity. Nat. Immunol. 2004;5:971–974. doi: 10.1038/ni1004-971. [DOI] [PubMed] [Google Scholar]
- Ip W.E., Hoshi N., Shouval D.S., Snapper S., Medzhitov R. Anti-inflammatory effect of IL-10 mediated by metabolic reprogramming of macrophages. Science. 2017;356:513–519. doi: 10.1126/science.aal3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin M.S., Kim S.E., Heo J.Y., Lee M.E., Kim H.M., Paik S.-G., Lee H., Lee J.-O. Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell. 2007;130:1071–1082. doi: 10.1016/j.cell.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Kawasaki T., Kawai T. Toll-like receptor signaling pathways. Front. Immunol. 2014;5:461. doi: 10.3389/fimmu.2014.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiu R., Hall L.J. An update on the human and animal enteric pathogen Clostridium perfringens. Emerg. Microbes Infect. 2018;7:1–15. doi: 10.1038/s41426-018-0144-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzeniewski C., Callewaert D.M. An enzyme-release assay for natural cytotoxicity. J. Immunol. Methods. 1983;64:313–320. doi: 10.1016/0022-1759(83)90438-6. [DOI] [PubMed] [Google Scholar]
- Lacey J.A., Johanesen P.A., Lyras D., Moore R.J. Genomic diversity of necrotic enteritis-associated strains of Clostridium perfringens: a review. Avian Pathol. 2016;45:302–307. doi: 10.1080/03079457.2016.1153799. [DOI] [PubMed] [Google Scholar]
- Laskin D.L. Macrophages and inflammatory mediators in chemical toxicity: a battle of forces. Chem. Res. Toxicol. 2009;22:1376–1385. doi: 10.1021/tx900086v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.S., Kim Y.-J. Signaling pathways downstream of pattern-recognition receptors and their cross talk. Annu. Rev. Biochem. 2007;76:447–480. doi: 10.1146/annurev.biochem.76.060605.122847. [DOI] [PubMed] [Google Scholar]
- Li Z., Wang W., Liu D., Guo Y. Effects of Lactobacillus acidophilus on the growth performance and intestinal health of broilers challenged with Clostridium perfringens. J. Anim. Sci. Biotechnol. 2018;9:25. doi: 10.1186/s40104-018-0243-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- M'Sadeq S.A., Wu S., Swick R.A., Choct M. Towards the control of necrotic enteritis in broiler chickens with in-feed antibiotics phasing-out worldwide. Anim. Nutr. 2015;1:1–11. doi: 10.1016/j.aninu.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y., Wang B., Xu X., Du W., Li W., Wang Y. Glycyrrhizic acid promotes M1 macrophage polarization in murine bone marrow-derived macrophages associated with the activation of JNK and NF-κB. Mediators Inflamm. 2015;2015:372931. doi: 10.1155/2015/372931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez F.O., Helming L., Gordon S. Alternative activation of macrophages: an immunologic functional perspective. Annu. Rev. Immunol. 2009;27:451–483. doi: 10.1146/annurev.immunol.021908.132532. [DOI] [PubMed] [Google Scholar]
- Mills C. M1 and M2 macrophages: oracles of health and disease. Crit. Rev. Immunol. 2012;32 doi: 10.1615/critrevimmunol.v32.i6.10. [DOI] [PubMed] [Google Scholar]
- Moore R.J. Necrotic enteritis predisposing factors in broiler chickens. Avian Pathol. 2016;45:275–281. doi: 10.1080/03079457.2016.1150587. [DOI] [PubMed] [Google Scholar]
- Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- Mosser D.M., Edwards J.P. Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray P.J. Macrophage polarization. Annu. Rev. Physiol. 2017;79:541–566. doi: 10.1146/annurev-physiol-022516-034339. [DOI] [PubMed] [Google Scholar]
- Nari N., Ghasemi H.A. Growth performance, nutrient digestibility, bone mineralization, and hormone profile in broilers fed with phosphorus-deficient diets supplemented with butyric acid and Saccharomyces boulardii. Poult. Sci. 2020;99:926–935. doi: 10.1016/j.psj.2019.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nerren J.R., He H., Genovese K., Kogut M.H. Expression of the avian-specific toll-like receptor 15 in chicken heterophils is mediated by gram-negative and gram-positive bacteria, but not TLR agonists. Vet. Immunol. Immunopathol. 2010;136:151–156. doi: 10.1016/j.vetimm.2010.02.017. [DOI] [PubMed] [Google Scholar]
- O'Brien D.K., Melville S.B. Effects of Clostridium perfringens alpha-toxin (PLC) and perfringolysin O (PFO) on cytotoxicity to macrophages, on escape from the phagosomes of macrophages, and on persistence of C. perfringens in host tissues. Infect. Immun. 2004;72:5204–5215. doi: 10.1128/IAI.72.9.5204-5215.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opal S.M., DePalo V.A. Anti-inflammatory cytokines. Chest. 2000;117:1162–1172. doi: 10.1378/chest.117.4.1162. [DOI] [PubMed] [Google Scholar]
- Palm N.W., Medzhitov R. Pattern recognition receptors and control of adaptive immunity. Immunol. Rev. 2009;227:221–233. doi: 10.1111/j.1600-065X.2008.00731.x. [DOI] [PubMed] [Google Scholar]
- Prescott J.F., Parreira V.R., Mehdizadeh Gohari I., Lepp D., Gong J. The pathogenesis of necrotic enteritis in chickens: what we know and what we need to know: a review. Avian Pathol. 2016;45:288–294. doi: 10.1080/03079457.2016.1139688. [DOI] [PubMed] [Google Scholar]
- Quezada S.A., Jarvinen L.Z., Lind E.F., Noelle R.J. CD40/CD154 interactions at the interface of tolerance and immunity. Annu. Rev. Immunol. 2004;22:307–328. doi: 10.1146/annurev.immunol.22.012703.104533. [DOI] [PubMed] [Google Scholar]
- Rajput I.R., Hussain A., Li Y.L., Zhang X., Xu X., Long M.Y., You D.Y., Li W.F. Saccharomyces boulardii and Bacillus subtilis B10 modulate TLRs mediated signaling to induce immunity by chicken BMDCs. J. Cell. Biochem. 2014;115:189–198. doi: 10.1002/jcb.24650. [DOI] [PubMed] [Google Scholar]
- Rajput I., Li L., Xin X., Wu B., Juan Z., Cui Z., Yu D., Li W. Effect of Saccharomyces boulardii and Bacillus subtilis B10 on intestinal ultrastructure modulation and mucosal immunity development mechanism in broiler chickens. Poult. Sci. 2013;92:956–965. doi: 10.3382/ps.2012-02845. [DOI] [PubMed] [Google Scholar]
- Ramlucken U., Ramchuran S.O., Moonsamy G., Lalloo R., Thantsha M.S., van Rensburg C.J. A novel Bacillus based multi-strain probiotic improves growth performance and intestinal properties of Clostridium perfringens challenged broilers. Poult. Sci. 2020;99:331–341. doi: 10.3382/ps/pez496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez B., Delgado S., Blanco-Míguez A., Lourenço A., Gueimonde M., Margolles A. Probiotics, gut microbiota, and their influence on host health and disease. Mol. Nutr. Food Res. 2017;61:1600240. doi: 10.1002/mnfr.201600240. [DOI] [PubMed] [Google Scholar]
- Schoster A., Kunz T., Lauper M., Graubner C., Schmitt S., Weese J. Prevalence of Clostridium difficile and Clostridium perfringens in Swiss horses with and without gastrointestinal disease and microbiota composition in relation to Clostridium difficile shedding. Vet. Microbiol. 2019;239:108433. doi: 10.1016/j.vetmic.2019.108433. [DOI] [PubMed] [Google Scholar]
- Shi H., Huang X., Yan Z., Yang Q., Wang P., Li S., Sun W., Gun S. Effect of Clostridium perfringens type C on TLR4/MyD88/NF-κB signaling pathway in piglet small intestines. Microb. Pathog. 2019;135:103567. doi: 10.1016/j.micpath.2019.103567. [DOI] [PubMed] [Google Scholar]
- Takehara M., Seike S., Sonobe Y., Bandou H., Yokoyama S., Takagishi T., Miyamoto K., Kobayashi K., Nagahama M. Clostridium perfringens α-toxin impairs granulocyte colony-stimulating factor receptor-mediated granulocyte production while triggering septic shock. Commun. Biol. 2019;2:1–12. doi: 10.1038/s42003-019-0280-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O., Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- Thomas S., Przesdzing I., Metzke D., Schmitz J., Radbruch A., Baumgart D.C. Saccharomyces boulardii inhibits lipopolysaccharide-induced activation of human dendritic cells and T cell proliferation. Clin. Exp. Immunol. 2009;156:78–87. doi: 10.1111/j.1365-2249.2009.03878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannella K.M., Wynn T.A. Mechanisms of organ injury and repair by macrophages. Annu. Rev. Physiol. 2017;79:593–617. doi: 10.1146/annurev-physiol-022516-034356. [DOI] [PubMed] [Google Scholar]
- Vergadi E., Ieronymaki E., Lyroni K., Vaporidi K., Tsatsanis C. Akt signaling pathway in macrophage activation and M1/M2 polarization. J. Immunol. 2017;198:1006–1014. doi: 10.4049/jimmunol.1601515. [DOI] [PubMed] [Google Scholar]
- Wade B., Keyburn A.L., Seemann T., Rood J.I., Moore R.J. Binding of Clostridium perfringens to collagen correlates with the ability to cause necrotic enteritis in chickens. Vet. Microbiol. 2015;180:299–303. doi: 10.1016/j.vetmic.2015.09.019. [DOI] [PubMed] [Google Scholar]
- Wang Y., Du W., Fu A., Zhang X., Huang Y., Lee K., Yu K., Li W., Li Y. Intestinal microbiota and oral administration of Enterococcus faecium associated with the growth performance of new-born piglets. Benef. Microbes. 2016;7:529–538. doi: 10.3920/BM2015.0099. [DOI] [PubMed] [Google Scholar]
- Wang B.-K., Mao Y.-L., Gong L., Xu X., Jiang S.-Q., Wang Y.-B., Li W.-F. Glycyrrhizic acid activates chicken macrophages and enhances their Salmonella-killing capacity in vitro. J. Zhejiang Univ. Sci. B. 2018;19:785–795. doi: 10.1631/jzus.B1700506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S.-B., Stanley D., Rodgers N., Swick R.A., Moore R.J. Two necrotic enteritis predisposing factors, dietary fishmeal and Eimeria infection, induce large changes in the caecal microbiota of broiler chickens. Vet. Microbiol. 2014;169:188–197. doi: 10.1016/j.vetmic.2014.01.007. [DOI] [PubMed] [Google Scholar]
- Wu Y., Wang Y., Zou H., Wang B., Sun Q., Fu A., Wang Y., Wang Y., Xu X., Li W. Probiotic Bacillus amyloliquefaciens SC06 induces autophagy to protect against pathogens in macrophages. Front. Microbiol. 2017;8:469. doi: 10.3389/fmicb.2017.00469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zotos D., Tarlinton D.M. Determining germinal centre B cell fate. Trends Immunol. 2012;33:281–288. doi: 10.1016/j.it.2012.04.003. [DOI] [PubMed] [Google Scholar]