FIGURE 4.
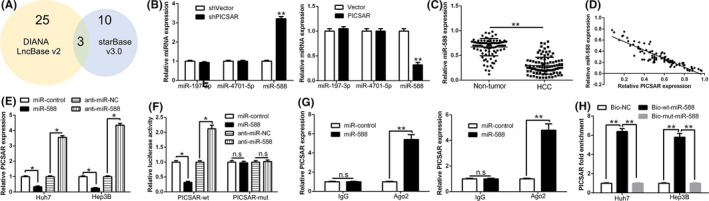
PICSAR functions as a sponge for microRNA (miR)‐588. A, LncBase and starBase were used to predict miRNAs that can bind to PICSAR. A total of three miRNAs, common to both databases, were identified. B, Quantitative RT‐PCR shows that miR‐588 expression was significantly upregulated among three selected miRNAs after PICSAR knockdown in Hep3B cells, whereas miR‐588 was downregulated after PICSAR overexpression in Huh7 cells. C, Expression of miR‐588 in tumor tissues was significantly lower than that in adjacent non‐tumor tissues. D, Pearson’s correlation analysis revealed a negative association between miR‐588 and PICSAR in hepatocellular carcinoma tissues. E, Expression of PICSAR was negatively regulated by miR‐588. F, Luciferase reporter gene assays showed that miR‐588 negatively regulated the luciferase activity of PICSAR‐WT‐3′‐UTR, rather than mutant PICSAR‐MUT‐3′‐UTR. G, Anti‐Ago2 RIPA with miR‐588 mimics showed that both miR‐588 and PICSAR were enriched in Ago2 precipitate compared to IgG. H, PICSAR was highly enriched in the sample pulled down by biotinylated WT miR‐588 rather than MUT miR‐588. n = three independent experiments. *P < .05, **P < .01
