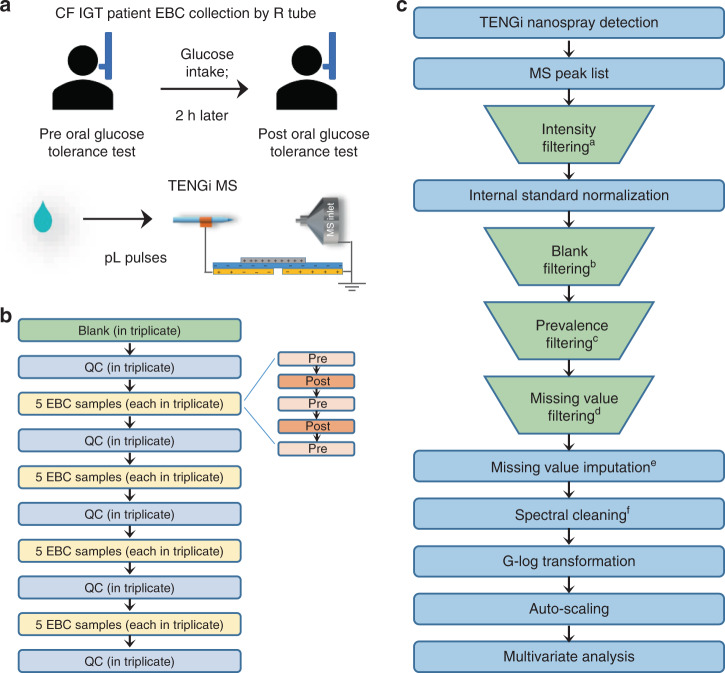
Fig. 4. Experimental and data processing workflow for TENGi MS EBC metabolomics.
a Illustration of EBC sample collection and TENGi MS analysis; b TENGi MS run order, including method blanks, quality control (QC) samples, and EBC sample replicates. Pre and Post samples were run in an interleaved fashion within each sample segment, and randomized in terms of their overall order. Three replicate tests were conducted for each sample. Outliers were discarded based on the extracted ion chronogram pulse area of the spiked IS; c TENGi MS data processing protocol. aSpectral features with abundances lower than 2000 were removed from the dataset. bBlank filtering was applied to remove features with normalized abundances lower than 3 times that of the blank. cA spectral feature had to be detected in at least 80% of the EBC samples, otherwise it was removed. dSpectral features in the QCs with 80% missing values or not detected were filtered out and sample(s) with more than 20% missing values were deemed as outlier(s) and removed (Supplementary Fig. 10). eMissing values were replaced by half of the minimum positive value in the original data. fQC features with median relative SD (RSDQC) > 30% were removed. The order of the four filtering processes is interchangeable.