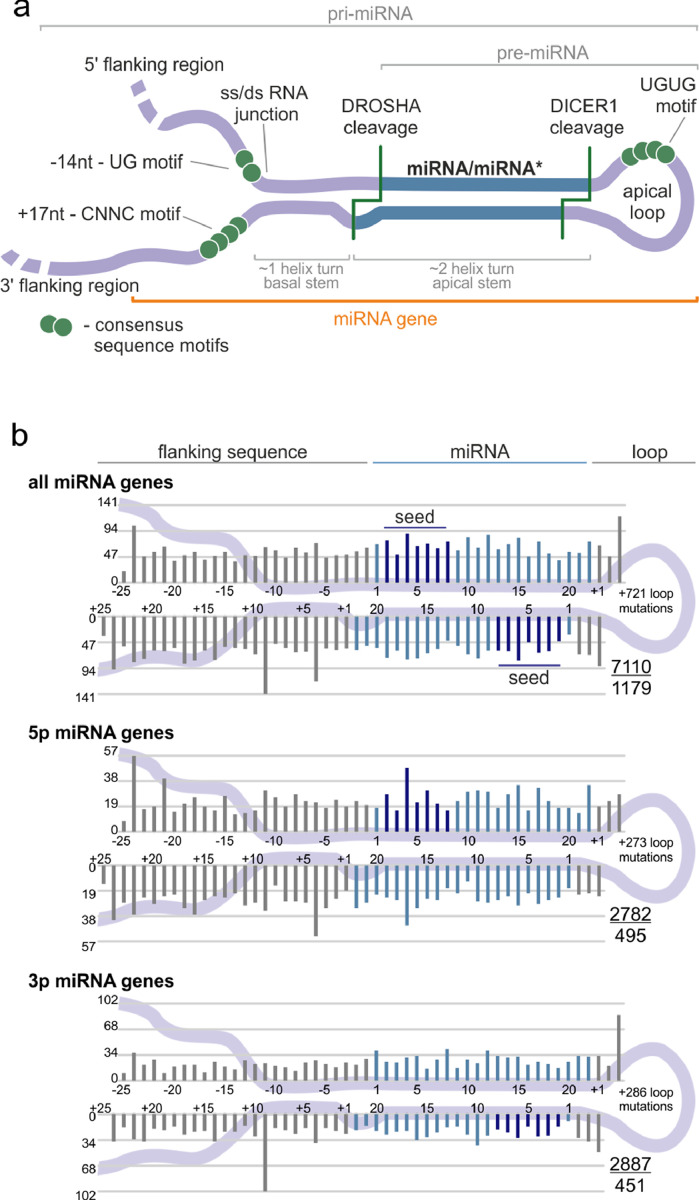
Fig. 2.
Localization of somatic mutations in miRNA precursors in Pan-Cancer. (a) An overview of a primary miRNA transcript with the indicated subregions considered in the study. The miRNA duplex is indicated in blue, and representative sequence consensus motifs recognized as enhancers of miRNA biogenesis are represented by green circles. (b) Localization of all detected mutations in the Pan-Cancer cohort. miRNA duplex positions are indicated in blue, seed regions in dark blue, and flanking regions and terminal positions of the apical loop in grey. The numbers in the lower-right corner represent the number of plotted mutations (upper) and the number of mutated miRNA genes (lower). If present, sequence variants localized beyond position 22 in longer mature miRNAs are shown cumulatively at position 22. The plot shows mutations within six positions of the loop (first 3 and last 3 nucleotides). The number of remaining mutations is indicated within the loop. Analyses were also performed in narrowed groups of miRNAs that release the guide miRNA strand predominantly from the 5p or 3p arm (lower panels).