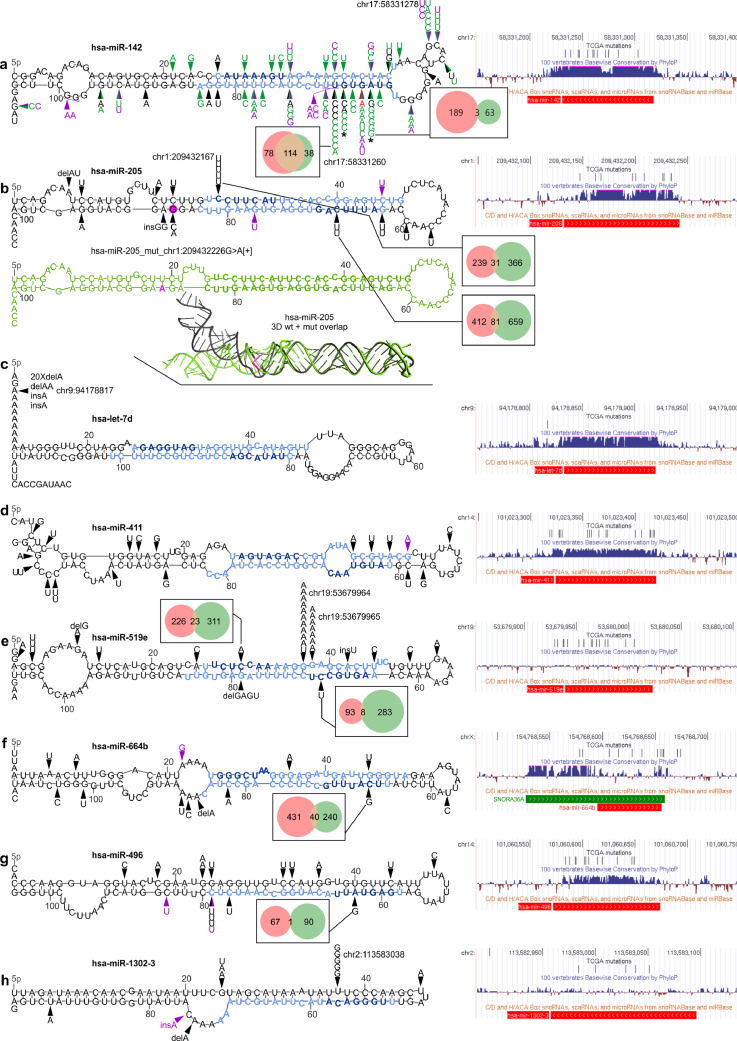
Fig. 4.
Localization of mutations in the selected overmutated miRNA genes. In each panel, on the left, mutations identified in the study in the TCGA samples (black arrowheads) and ICGC-PCAWG samples (purple arrowheads) are shown on mfold-predicted 2D structures of the miRNA precursors. Mature miRNA and seed sequences are indicated in light blue and dark blue, respectively. On the right, a screenshots from the UCSC genome browser, showing (from the top) a custom track with positions of the TCGA mutations detected in the study, conservation of the genomic region, and position of short non-coding RNAs, including pre-miRNAs (red bar; according to miRbase) and snoRNAs (green bar). For selected seed mutations, Venn-diagrams indicate the number of predicted targets for the wild-type (pink circle) and mutated (green circle) seeds. The effect of the seed mutations on the levels of the selected mutation-specific targets is shown in Supplementary Fig. S4. Genomic positions of the most significant hotspots are indicated next to mutations symbol. (a) Mutations found in hsa-miR-142. Green arrowheads indicate mutations detected in previous studies [[57], [58], [59], [60], [61],[63], [64], [65]] (for details see Supplementary Table S8); red arrowhead indicates the mutation identified by us in the Burkitt's lymphoma (Raji) cell line. * indicates the mutations tested functionally [27]. (b) Mutations found in hsa-miR-205 (above). Below: (i) the corresponding 2D structure of the precursor with the chr1:209432226G>A[+] mutation (drawn in green) and (ii) superimposed 3D structures of the wild-type (black) and mutant (green) precursors are shown. The position of the mutation and the mutant allele are marked in pink. (c-h) Mutations found in hsa-let-7d, hsa-miR-411, hsa-miR-519e, hsa-miR-664b, hsa-miR-496, and hsa-miR-1302-3, respectively.