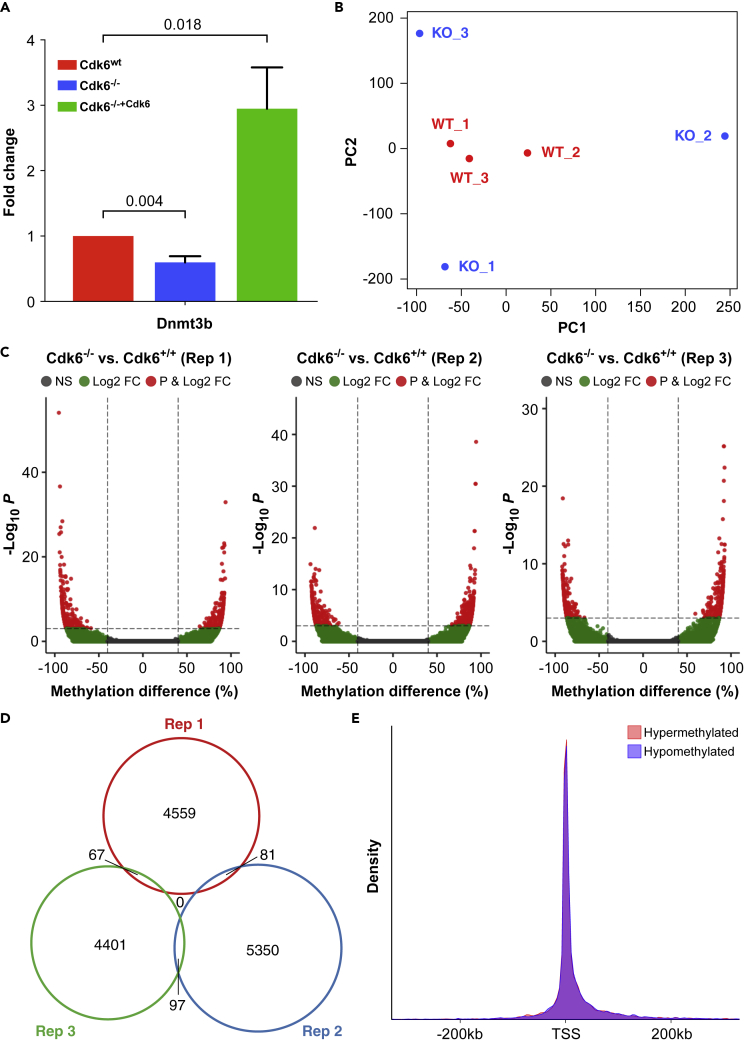
Figure 3.
Analysis of the Methylome of BCR-ABL+ Cdk6wt and BCR-ABL+ Cdk6−/− Cells By Reduced Representation Bisulfite Sequencing
(A) Quantification of Dnmt3b mRNA expression in BCR-ABL+ Cdk6wt, BCR-ABL+ Cdk6−/− and BCR-ABL+ Cdk6−/−+Cdk6 cells determined by RT-PCR. Mean fold changes +SEM are shown (N = 8), p values were calculated using one-sample t-tests.
(B) Principal component analysis of 3 BCR-ABL+ Cdk6wt (WT, red) and 3 BCR-ABL+ Cdk6−/− (KO, blue) cell lines based on CpG methylation. Each dot represents a unique sample. See also Figure S4.
(C) Volcano plots showing hypomethylated and hypermethylated CpG sites in 3 replicates of BCR-ABL+ Cdk6−/− cell lines compared to BCR-ABL+ Cdk6wt cells.
(D) Venn diagram illustrating the overlap of differentially methylated CpG sites in the 3 BCR-ABL+ Cdk6−/− cell lines.
(E) Density plot showing the location of hypermethylated (pink) and hypomethylated (blue) CpG sites relative to transcriptional start sites (TSS). See also Figure S4.