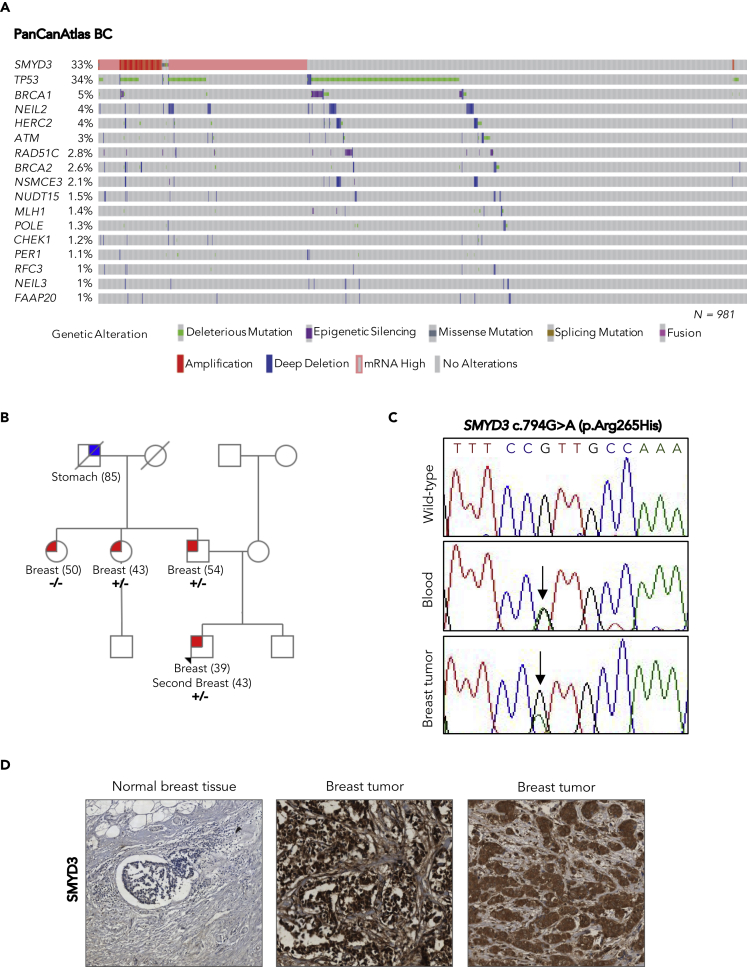
Figure 3.
SMYD3 Analysis in the PanCanAtlas BC Dataset and Identification of a Genetic Variant in a High-Risk BC Family
(A) Oncoprint of SMYD3 and HRD-associated genes in PanCanAtlas BC tumors. Overall profiling of 981 BC tumors (columns) carrying alterations involving the SMYD3 gene (mRNA overexpression, copy number alterations, and mutations) and deleterious mutations, deletions, and epigenetic silencing events for each HRD-associated gene (rows with gene names listed on the left) in more than 1% of cases. Gray boxes indicate the absence of alterations, and color/shape combinations corresponding to the various alteration types are indicated below the oncoprint. The overall frequency of each gene alteration in the oncoprint plot is indicated on the left.
(B) Pedigree of the BC family selected for whole-exome sequencing. The proband is indicated by an arrowhead. For each individual diagnosed with cancer, the age at diagnosis is reported. Tested family members are marked as +/− to indicate SMYD3 (C)794G>A (p.Arg265His) heterozygous mutation carriers or as −/− to indicate wild-type SMYD3 individuals.
(C) Partial electropherogram of SMYD3 exon 8 confirming the presence of the SMYD3 c.794G>A mutation (indicated by a black arrow) in both the germline and the tumor DNA of the proband.
(D) SMYD3 expression by immunohistochemistry in normal male breast tissue (left) and male breast tumors: cytoplasmic only localization (center) and nuclear and cytoplasmic localization (right). All the analyzed BC sections showed strong (3+) SMYD3 expression compared with normal breast tissue, regardless of SMYD3 mutational status. Magnification: 20×.
Results are representative of at least three independent experiments. See also Figure S4 and Table S1.