Abstract
The Genetics Society of America’s (GSA’s) Edward Novitski Prize recognizes a single experimental accomplishment or a body of work in which an exceptional level of creativity, and intellectual ingenuity, has been used to design and execute scientific experiments to solve a difficult problem in genetics. The 2020 recipient is Welcome W. Bender of Harvard Medical School, recognizing his creativity and ingenuity in revealing the molecular nature and regulation of the bithorax gene complex.

Of all of the so-called complex loci explored by Drosophila geneticists, the bithorax complex (BX-C) stands out as most extensive and intriguing. That is thanks to decades of study by E.B. Lewis and his colleagues, summarized in his 1995 Nobel lecture “The Bithorax Complex: the First Fifty Years.” When techniques were developed to isolate Drosophila loci based on their chromosomal position, the bithorax complex was the most appealing target. The renewed description of genetic observations in molecular terms has led to insights widely applicable in molecular biology. This is an attempt to highlight the most important of those insights.
The initial recovery of molecular clones covering the bithorax complex, and mapping of mutant lesions, confirmed Lewis’ observations, based on classical genetic recombination, that the locus was large (over 300 kb), and that the chromosomal order of mutations aligned with the order of the body segments most affected (Bender et al. 1983a,b; Karch et al. 1985) (see Figure 1). What was the nature of all of this DNA? Contemporaneous saturation mutagenesis suggested that the BX-C probably contained only three lethal complementation groups (Sánchez-Herrero et al. 1985; Tiong et al. 1985), but it was still surprising that so few protein coding transcripts were found, principally those for Ultrabithorax (O’Connor et al. 1988; Kornfeld et al. 1989), abdominal-A (Karch et al. 1990), and Abdominal-B (Kuziora and McGinnis 1988; Celniker et al. 1989; Zavortink and Sakonju 1989) (see Figure 1). The comparison of the predicted proteins for Ultrabithorax, and for another segment identity gene, Antennapedia, led to the discovery of the homeobox (McGinnis et al. 1984a; Scott and Weiner 1984)—a DNA binding motif widely used in developmental regulation. These segment identity homeobox genes were strikingly conserved; homologous “HOX” clusters were discovered in many animals, including mammals (McGinnis et al. 1984b; Akam 1989). The recognition of this functional conservation suddenly elevated the importance of Drosophila as a model for human biology.
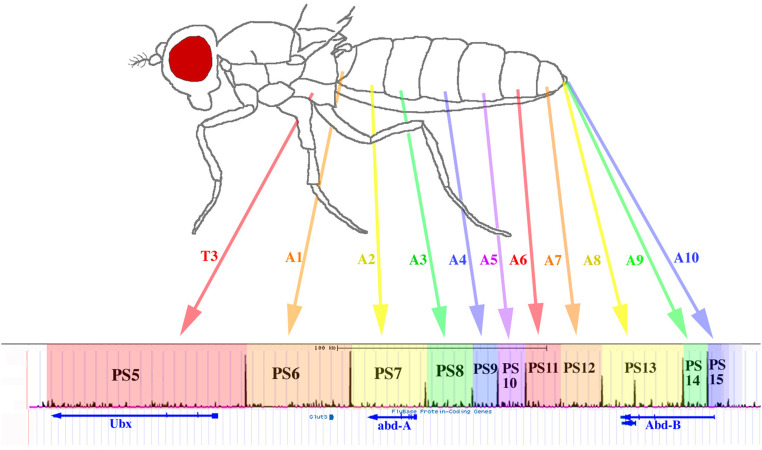
Figure 1.
Regulatory Domains of the bithorax complex. A diagram of an adult female fly shows the body segments specified by the BX-C, including the third thoracic (T3) and 10 abdominal segments (A1–A10). The 8th, 9th, and 10th abdominal segments contribute only small rudiments. The map below shows the embryonic binding sites of the CTCF protein along the 325 kb extent of the BX-C (Bowman et al. 2014). At the bottom are shown the transcription units of the three homeobox genes, Ubx, abd-A, and Abd-B. The colored blocks of the map show 11 successive regulatory domains, marked with parasegment numbers (PS5-15). The arrows connect the body segments with their corresponding parasegmental domains. Most domain boundaries correspond to CTCF binding sites. The righthand boundaries of domains PS5-7 and PS11-14 have been confirmed by mapping the extent of histone modifications (H3K27me3) in nuclei from single parasegments (Bowman et al. 2014; S. Tabor, H. Domingues, and W. Bender, unpublished data).
The dearth of proteins encoded in the BX-C suggested that the vast majority of the sequence, most of which is evolutionarily conserved, must be regulatory. That the regulation of a transcription factor could be so intricate was a novel realization, but it made sense when the expression patterns of the Hox proteins were found to be exceptionally granular in time and space (White and Wilcox 1984; Beachy et al. 1985; Kuziora and McGinnis 1988; Celniker et al. 1989; Delorenzi and Bienz 1990; Macias et al. 1990; Karch et al. 1990; Boulet et al. 1991; Sánchez-Herrero 1991).
A pleasant surprise of the early mapping was that mutant lesions were so easy to find. Point mutations outside of the coding regions were very rare, despite multiple mutagenesis screens by Lewis using the chemical mutagen ethyl methane sulfonate (EMS). Regulatory sequences, such as enhancers, are difficult to damage with single base changes. X-rays, by contrast, generated several deletions and many rearrangement breaks; mapping these made it possible to discern the layout of the genes of the BX-C and their regulatory regions.
Most spontaneous mutations in the BX-C and elsewhere were associated with mobile element insertions. Curiously, the majority of insertions in the BX-C with penetrant phenotypes were due to one particular transposable element named “gypsy.” These and other gypsy-induced mutations could be phenotypically reverted by a second-site mutation in the gene for suppressor-of-Hairy-wing [su(Hw)] (Modolell et al. 1983). Some revertants of gypsy-induced mutations had small deletions within gypsy in a region of tandem repeats (Peifer and Bender 1988), shown to be binding sites for the SU(HW) protein, a zinc-finger DNA-binding protein (Spana et al. 1988). The cluster of SU(HW) binding sites in the gypsy element was an early example of an “insulator,” able to block enhancer/promoter interactions (Geyer and Corces 1992; Gdula et al. 1996).
Ed Lewis highlighted the sequential arrangement of his mutations on the genetic map with the anterior/posterior order of the body segments they most affect—his “colinearity rule.” He suggested a separate functional region for each segment, from the third thoracic to the eighth abdominal (Lewis 1981). His model was repeated with more molecular wording, with a cis-regulatory “domain” for each segment (Peifer et al. 1987), or, more properly, for each parasegment (Martinez-Arias and Lawrence 1985). The central idea was that individual regulatory domains were only “open for business” in specific body segments (Maeda and Karch 2015). This idea was reinforced when P element reporter constructs were recovered that were inserted within the BX-C. Their reporter proteins (LacZ, GFP, or Gal4) were expressed only in the segments specific to the domain where the insertion landed (McCall et al. 1994; Bender and Hudson 2000).
The features of a BX-C domain have been gradually elaborated. The anterior/posterior positional cues for each domain were presumed to come from the “gap” and “pair rule” genes that lay down the segments within the first few hours of embryonic development (Nüsslein-Volhard and Wieschaus 1980). Candidate “embryonic enhancers” or “initiators” were first identified in P element transgenes carrying various restriction fragments from the BX-C (Simon et al. 1990; Mihaly et al. 2006). Their positions were refined and correlated with binding sites for gap and pair rule genes (Qian et al. 1993), and their relevance was confirmed by the discovery of rare single-base-change mutations in gap gene binding sites in these enhancers (Shimell et al. 1994; Ho et al. 2009). The most definitive demonstration involved the replacement within the BX-C of the initiator for the sixth abdominal segment with that of the fifth; the developmental program for the sixth abdominal segment was turned on ectopically in the fifth abdominal segment (Iampietro et al. 2010). There are many other enhancers within each domain, first recognized in transgenes (Pirrotta et al. 1995), which specify the cell types where the homeotic genes are expressed.
Boundaries between BX-C regulatory domains were recognized after a striking class of dominant mutations was linked to small deletions. In the prototypic example, Fab-7, a 4 kb deletion between the domains for the sixth and seventh abdominal segments, transformed the sixth segment into a copy of the seventh (Gyurkovics et al. 1990). In the fused domain, the developmental instructions for the seventh abdominal segment came under the control of the positional cues for the sixth abdominal segment. The BX-C boundaries are sites of binding by the CTCF protein, another zinc-finger DNA-binding protein (Holohan et al. 2007) (see Figure 1). Many proposed boundaries in mammalian systems are also marked by CTCF, but the function of a boundary is still best demonstrated by the examples in the BX-C.
The third critical element of each domain is a “Polycomb Response Element” (PRE, Simon et al. 1993). Lewis showed that the BX-C is negatively regulated by the Polycomb gene (Lewis 1978), and studies of BX-C protein patterns showed that the segmentally limited expression in wild type embryos spread to all segments in Polycomb mutants (Beachy et al. 1985; Celniker et al. 1989; Simon et al. 1992). The POLYCOMB protein was found bound to several discrete sites within the BX-C (Chiang et al. 1995; Schwartz et al. 2006). A few of these PREs have been dissected and shown to include binding sites for Polycomb and for other gene products of the Polycomb Group (Horard et al. 2000; Sipos et al. 2007; Orsi et al. 2014). Because repression by the Polycomb Group acted on a variety of reporter constructs introduced into the BX-C, such repression seemed likely to involve a blockage to accessibility by transcription factors or polymerase. Restricted accessibility was confirmed by in vivo assays (Fitzgerald and Bender 2001), and was recently corroborated by chromatin compaction of repressed domains, as seen in super-resolution microscopy (Boettiger et al. 2016; Mateo et al. 2019). Polycomb repression is accompanied by trimethylation of lysine 27 of histone H3 (Müller et al. 2002). Assays of nuclei from single segments show loss of H3K27me3 correlates with the sequential activation of each domain (Bowman et al. 2014).
The domain model is perhaps the most important contribution from molecular studies of the BX-C. Each domain, with an initiator, a PRE, and flanking boundaries, is a subroutine in a larger program of development, with instructions reserved to be read only at a particular time and place. Genome-wide surveys of H3K27me3 show many other loci with broad regions of methylation (Schwartz et al. 2006); the number of loci with this sort of domain architecture will likely increase as more tissues and times are assayed.
Many outstanding questions in gene regulation and nuclear architecture might be best answered in future studies of the BX-C. We have no idea how initiator elements talk to PREs to impose or block repression. We know of many “noncoding” RNAs transcribed from within the BX-C (Lipshitz et al. 1987; Sánchez-Herrero and Akam 1989; Graveley et al. 2011; Pease et al. 2013), but, with rare exceptions (Gummalla et al. 2012; Maeda et al. 2018), we know nothing of their functions. We know that enhancers must act on distant promoters, even on opposite chromosomes (“transvection”; Lewis 1954, Duncan 2002), but aside from vague notions of “looping,” we do not know how target promoters are found. Domain boundaries with CTCF attached can sometimes be ignored or “bypassed” (Kyrchanova et al. 2019), but we do not know how this is regulated. Perhaps the most glaring curtain of ignorance surrounds Lewis’ initial rule of colinearity; why must the domains be lined up on the chromosome in the order of the body segments? The well is far from dry.
A current trend in molecular biology is to do genome-wide investigations, with results often distilled into a “metagene.” This serves to establish the generality of known mechanisms. A more traditional approach is to focus on a single locus or system, preferably one with a rich legacy of genetic description. This is arguably the more promising path to discovery of novel mechanisms. To the textbook examples of the Lac operon, phage lambda, and yeast mating type, perhaps we can add the Drosophila BX-C.
Acknowledgments
Thoughtful improvements to this manuscript were provided by François Karch and Mark Peifer. Long-term funding for our work on the BX-C has been provided by the National Institutes of Health.
Literature Cited
- Akam M., 1989. Hox and HOM: homologous gene clusters in insects and vertebrates. Cell 57: 347–349. 10.1016/0092-8674(89)90909-4 [DOI] [PubMed] [Google Scholar]
- Beachy P. A., Helfand S. L., and Hogness D. S., 1985. Segmental distribution of bithorax complex proteins during Drosophila development. Nature 313: 545–551. 10.1038/313545a0 [DOI] [PubMed] [Google Scholar]
- Bender W., and Hudson A., 2000. P element homing to the Drosophila bithorax complex. Development 127: 3981–3992. [DOI] [PubMed] [Google Scholar]
- Bender W., Spierer P., and Hogness D. S., 1983a Chromosomal walking and jumping to isolate DNA from the Ace and rosy loci and the bithorax complex in Drosophila melanogaster. J. Mol. Biol. 168: 17–33. 10.1016/S0022-2836(83)80320-9 [DOI] [PubMed] [Google Scholar]
- Bender W., Akam M., Karch F., Beachy P. A., Peifer M. et al. , 1983b Molecular genetics of the bithorax complex in Drosophila melanogaster. Science 221: 23–29. 10.1126/science.221.4605.23 [DOI] [PubMed] [Google Scholar]
- Boettiger A. N., Bintu B., Moffitt J. R., Wang S., Beliveau B. J. et al. , 2016. Super-resolution imaging reveals distinct chromatin folding for different epigenetic states. Nature 529: 418–422. 10.1038/nature16496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulet A. M., Lloyd A., and Sakonju S., 1991. Molecular definition of the morphogenetic and regulatory functions and the cis-regulatory elements of the Drosophila Abd-B homeotic gene. Development 111: 393–405. [DOI] [PubMed] [Google Scholar]
- Bowman S. K., Deaton A. M., Domingues H., Wang P. I., Sadreyev R. I. et al. , 2014. H3K27 modifications define segmental regulatory domains in the Drosophila bithorax complex. eLife 3: e02833 10.7554/eLife.02833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker S. E., Keelan D. J., and Lewis E. B., 1989. The molecular genetics of the bithorax complex of Drosophila: characterization of the products of the abdominal-B domain. Genes Dev. 3: 1424–1436. 10.1101/gad.3.9.1424 [DOI] [PubMed] [Google Scholar]
- Chiang A., O’Connor M. B., Paro R., Simon J., and Bender W., 1995. Discrete polycomb-binding sites in each parasegmental domain of the bithorax complex. Development 121: 1681–1689. [DOI] [PubMed] [Google Scholar]
- Delorenzi M., and Bienz M., 1990. Expression of abdominal-B homeoproteins in Drosophila embryos. Development 108: 323–329. [DOI] [PubMed] [Google Scholar]
- Duncan I. W., 2002. Transvection effects in Drosophila. Annu. Rev. Genet. 36: 521–556. 10.1146/annurev.genet.36.060402.100441 [DOI] [PubMed] [Google Scholar]
- Fitzgerald D. P., and Bender W., 2001. Polycomb group repression reduces DNA accessibility. Mol. Cell. Biol. 21: 6585–6597. 10.1128/MCB.21.19.6585-6597.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gdula D. A., Gerasimova T. I., and Corces V. G., 1996. Genetic and molecular analysis of the gypsy chromatin insulator of Drosophila. Proc. Natl. Acad. Sci. USA 93: 9378–9383. 10.1073/pnas.93.18.9378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer P. K., and Corces V. G., 1992. DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev. 6: 1865–1873. 10.1101/gad.6.10.1865 [DOI] [PubMed] [Google Scholar]
- Graveley B. R., Brooks A. N., Carlson J. W., Duff M. O., Landolin J. M. et al. , 2011. The developmental transcriptome of Drosophila melanogaster. Nature 471: 473–479. 10.1038/nature09715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gummalla M., Maeda R. K., Castro Alvarez J. J., Gyurkovics H., Singari S. et al. , 2012. abd-A regulation by the iab-8 noncoding RNA. PLoS Genet. 8: e1002720 10.1371/journal.pgen.1002720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyurkovics H., Gausz J., Kummer J., and Karch F., 1990. A new homeotic mutation in the Drosophila bithorax complex removes a boundary separating two domains of regulation. EMBO J. 9: 2579–2585. 10.1002/j.1460-2075.1990.tb07439.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho M. C. W., Johnsen H., Goetz S. E., Schiller B. J., Bae E. et al. , 2009. Functional evolution of cis-regulatory modules at a homeotic gene in Drosophila. PLoS Genet. 5: e1000709 10.1371/journal.pgen.1000709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holohan E. E., Kwong C., Adryan B., Bartkuhn M., Herold M. et al. , 2007. CTCF genomic binding sites in Drosophila and the organisation of the bithorax complex. PLoS Genet. 3: e112 10.1371/journal.pgen.0030112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horard B., Tatout C., Poux S., and Pirrotta V., 2000. Structure of a polycomb response element and in vitro binding of polycomb group complexes containing GAGA factor. Mol. Cell. Biol. 20: 3187–3197. 10.1128/MCB.20.9.3187-3197.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iampietro C., Gummalla M., Mutero A., Karch F., and Maeda R. K., 2010. Initiator elements function to determine the activity state of BX-C enhancers. PLoS Genet. 6: e1001260 10.1371/journal.pgen.1001260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karch F., Weiffenbach B., Peifer M., Bender W., Duncan I. et al. , 1985. The abdominal region of the bithorax complex. Cell 43: 81–96. 10.1016/0092-8674(85)90014-5 [DOI] [PubMed] [Google Scholar]
- Karch F., Bender W., and Weiffenbach B., 1990. abdA expression in Drosophila embryos. Genes Dev. 4: 1573–1587. 10.1101/gad.4.9.1573 [DOI] [PubMed] [Google Scholar]
- Kornfeld K., Saint R. B., Beachy P. A., Harte P. J., Peattie D. A. et al. , 1989. Structure and expression of a family of Ultrabithorax mRNAs generated by alternative splicing and polyadenylation in Drosophila. Genes Dev. 3: 243–258. 10.1101/gad.3.2.243 [DOI] [PubMed] [Google Scholar]
- Kuziora M. A., and McGinnis W., 1988. Different transcripts of the Drosophila Abd-B gene correlate with distinct genetic sub-functions. EMBO J. 7: 3233–3244. 10.1002/j.1460-2075.1988.tb03190.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrchanova O., Sabirov M., Mogila V., Kurbidaeva A., Postika N. et al. , 2019. Complete reconstitution of bypass and blocking functions in a minimal artificial Fab-7 insulator from Drosophila bithorax complex. Proc. Natl. Acad. Sci. USA 116: 13462–13467. 10.1073/pnas.1907190116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis E. B., 1954. The theory and application of a new method of detecting chromosomal rearrangements in Drosophila melanogaster. Am. Nat. 88: 225–239. 10.1086/281833 [DOI] [Google Scholar]
- Lewis E. B., 1978. A gene complex controlling segmentation in Drosophila. Nature 276: 565–570. 10.1038/276565a0 [DOI] [PubMed] [Google Scholar]
- Lewis E. B., 1981. Developmental biology using purified genes, pp. 189–208 in ICN-UCLA Symposia on Molecular and Cellular Biology, Vol. 23, edited by Brown D. D. and Fox C. F.. Academic, New York. [Google Scholar]
- Lipshitz H. D., Peattie D. A., and Hogness D. S., 1987. Novel transcripts from the Ultrabithorax domain of the bithorax complex. Genes Dev. 1: 307–322. 10.1101/gad.1.3.307 [DOI] [PubMed] [Google Scholar]
- Macias A., Casanova J., and Morata G., 1990. Expression and regulation of the abd-A gene of Drosophila. Development 110: 1197–1207. [DOI] [PubMed] [Google Scholar]
- Maeda R. K., and Karch F., 2015. The open for business model of the bithorax complex in Drosophila. Chromosoma 124: 293–307. 10.1007/s00412-015-0522-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda R. K., Sitnik J. L., Frei Y., Prince E., Gligorov D. et al. , 2018. The lncRNA male-specific abdominal plays a critical role in Drosophila accessory gland development and male fertility. PLoS Genet. 14: e1007519 10.1371/journal.pgen.1007519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Arias A., and Lawrence P. A., 1985. Parasegments and compartments in the Drosophila embryo. Nature 313: 639–642. 10.1038/313639a0 [DOI] [PubMed] [Google Scholar]
- Mateo L. J., Murphy S. E., Hafner A., Cinquini I. S., Walker C. A. et al. , 2019. Visualizing DNA folding and RNA in embryos at single-cell resolution. Nature 568: 49–54. 10.1038/s41586-019-1035-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCall K., O’Connor M. B., and Bender W., 1994. Enhancer traps in the Drosophila bithorax complex mark parasegmental domains. Genetics 138: 387–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis W., Levine M. S., Hafen E., Kuroiwa A., and Gehring W. J., 1984a A conserved DNA sequence in homoeotic genes of the Drosophila Antennapedia and bithorax complexes. Nature 308: 428–433. 10.1038/308428a0 [DOI] [PubMed] [Google Scholar]
- McGinnis W., Garber R. L., Wirz J., Kuroiwa A., and Gehring W. J., 1984b A homologous protein-coding sequence in Drosophila homeotic genes and its conservation in other metazoans. Cell 37: 403–408. 10.1016/0092-8674(84)90370-2 [DOI] [PubMed] [Google Scholar]
- Mihaly J., Barges S., Sipos L., Maeda R., Cléard F. et al. , 2006. Dissecting the regulatory landscape of the Abd-B gene of the bithorax complex. Development 133: 2983–2993. 10.1242/dev.02451 [DOI] [PubMed] [Google Scholar]
- Modolell J., Bender W., and Meselson M., 1983. Drosophila melanogaster mutations suppressible by the suppressor of Hairy-wing are insertions of a 7.3-kilobase mobile element. Proc. Natl. Acad. Sci. USA 80: 1678–1682. 10.1073/pnas.80.6.1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J., Hart C. M., Francis N. J., Vargas M. L., Sengupta A. et al. , 2002. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111: 197–208. 10.1016/S0092-8674(02)00976-5 [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C., and Wieschaus E., 1980. Mutations affecting segment number and polarity in Drosophila. Nature 287: 795–801. 10.1038/287795a0 [DOI] [PubMed] [Google Scholar]
- O’Connor M. B., Binari R., Perkins L. A., and Bender W., 1988. Alternative RNA products from the Ultrabithorax domain of the bithorax complex. EMBO J. 7: 435–445. 10.1002/j.1460-2075.1988.tb02831.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsi G. A., Kasinathan S., Hughes K. T., Saminadin-Peter S., Henikoff S. et al. , 2014. High-resolution mapping defines the cooperative architecture of Polycomb response elements. Genome Res. 24: 809–820. 10.1101/gr.163642.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pease B., Borges A. C., and Bender W., 2013. Noncoding RNAs of the Ultrabithorax domain of the Drosophila bithorax complex. Genetics 195: 1253–1264. 10.1534/genetics.113.155036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peifer M., and Bender W., 1988. Sequences of the gypsy transposon of Drosophila necessary for its effects on adjacent genes. Proc. Natl. Acad. Sci. USA 85: 9650–9654. 10.1073/pnas.85.24.9650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peifer M., Karch F., and Bender W., 1987. The bithorax complex: control of segmental identity. Genes Dev. 1: 891–898. 10.1101/gad.1.9.891 [DOI] [PubMed] [Google Scholar]
- Pirrotta V., Chan C. S., McCabe D., and Qian S., 1995. Distinct parasegmental and imaginal enhancers and the establishment of the expression pattern of the Ubx gene. Genetics 141: 1439–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian S., Capovilla M., and Pirrotta V., 1993. Molecular mechanisms of pattern formation by the BRE enhancer of the Ubx gene. EMBO J. 12: 3865–3877. 10.1002/j.1460-2075.1993.tb06065.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Herrero E., 1991. Control of the expression of the bithorax complex genes abdominal-A and abdominal-B by cis-regulatory regions in Drosophila embryos. Development 111: 437–449. [DOI] [PubMed] [Google Scholar]
- Sánchez-Herrero E., and Akam M., 1989. Spatially ordered transcription of regulatory DNA in the bithorax complex of Drosophila. Development 107: 321–329. [DOI] [PubMed] [Google Scholar]
- Sánchez-Herrero E., Vernós I., Marco R., and Morata G., 1985. Genetic organization of Drosophila bithorax complex. Nature 313: 108–113. 10.1038/313108a0 [DOI] [PubMed] [Google Scholar]
- Schwartz Y. B., Kahn T. G., Nix D. A., Li X.-Y., Bourgon R. et al. , 2006. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat. Genet. 38: 700–705. 10.1038/ng1817 [DOI] [PubMed] [Google Scholar]
- Scott M. P., and Weiner A. J., 1984. Structural relationships among genes that control development: sequence homology between the Antennapedia, Ultrabithorax, and fushi tarazu loci of Drosophila. Proc. Natl. Acad. Sci. USA 81: 4115–4119. 10.1073/pnas.81.13.4115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimell M. J., Simon J., Bender W., and O’Connor M. B., 1994. Enhancer point mutation results in a homeotic transformation in Drosophila. Science 264: 968–971. 10.1126/science.7909957 [DOI] [PubMed] [Google Scholar]
- Simon J., Peifer M., Bender W., and O’Connor M., 1990. Regulatory elements of the bithorax complex that control expression along the anterior-posterior axis. EMBO J. 9: 3945–3956. 10.1002/j.1460-2075.1990.tb07615.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon J., Chiang A., and Bender W., 1992. Ten different Polycomb group genes are required for spatial control of the abdA and AbdB homeotic products. Development 114: 493–505. [DOI] [PubMed] [Google Scholar]
- Simon J., Chiang A., Bender W., Shimell M. J., and O’Connor M., 1993. Elements of the Drosophila bithorax complex that mediate repression by Polycomb group products. Dev. Biol. 158: 131–144. 10.1006/dbio.1993.1174 [DOI] [PubMed] [Google Scholar]
- Sipos L., Kozma G., Molnár E., and Bender W., 2007. In situ dissection of a Polycomb response element in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 104: 12416–12421. 10.1073/pnas.0703144104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spana C., Harrison D. A., and Corces V. G., 1988. The Drosophila melanogaster suppressor of Hairy-wing protein binds to specific sequences of the gypsy retrotransposon. Genes Dev. 2: 1414–1423. 10.1101/gad.2.11.1414 [DOI] [PubMed] [Google Scholar]
- Tiong S., Bone L. M., and Whittle J. R., 1985. Recessive lethal mutations within the bithorax-complex in Drosophila. Mol. Gen. Genet. 200: 335–342. 10.1007/BF00425445 [DOI] [PubMed] [Google Scholar]
- White R. A., and Wilcox M., 1984. Protein products of the bithorax complex in Drosophila. Cell 39: 163–171. 10.1016/0092-8674(84)90202-2 [DOI] [PubMed] [Google Scholar]
- Zavortink M., and Sakonju S., 1989. The morphogenetic and regulatory functions of the Drosophila Abdominal-B gene are encoded in overlapping RNAs transcribed from separate promoters. Genes Dev. 3: 1969–1981. 10.1101/gad.3.12a.1969 [DOI] [PubMed] [Google Scholar]