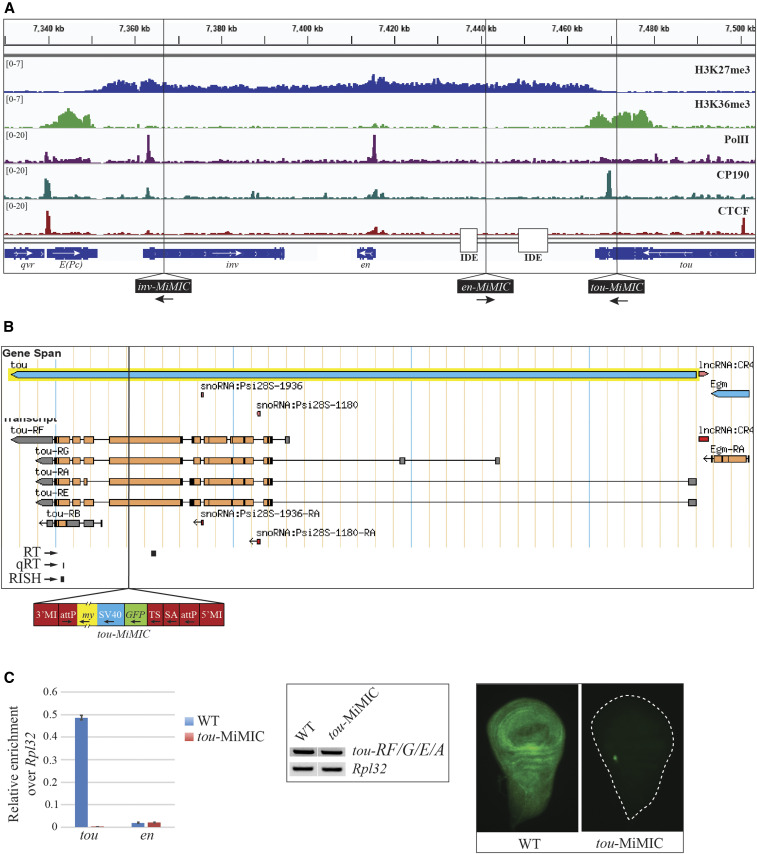
Figure 2.
tou-MiMIC stops tou transcription. (A) ChIP-seq distribution of H3K27me3, H3K36me3, PolII, CP190, and CTCF over the inv-en domain in WT. ChIP-seq data shown here and throughout this study are derived from brains and discs from third-instar larvae. H3K27me3 covers the entire inv-en domain, from the 3′ end of E(Pc) to the 3′ end of tou. In contrast, H3K36me3, a mark of actively transcribed genes, is strong over E(Pc) and tou but undetectable over the inv and en transcription units. Vertical black lines show the insertion site of the three MiMIC transgenes used in this study (bottom). The locations of the imaginal disc enhancers (IDE) are also shown. (B) tou gene and annotated transcript isoforms from FlyBase with the tou-MiMIC insertion site indicated. Positions of the tou qRT-PCR amplicon (qRT), the tou RT-PCR amplicon (RT), and the tou-all in situ probe (RISH) used in this figure are shown below the tou isoforms. The MiMIC element is diagrammed below (my: mini-yellow, SV: SV40 terminator, TS: translation stop, SA: splice acceptor), (C) Left panel: qRT-PCR analysis of tou and en transcripts in WT and tou-MiMIC. Middle panel: RT-PCR analysis of tou-RF/G/E/A, designed to amplify all tou transcripts except tou-RB. Rpl32 amplicons were used as a loading control. Right panel: RISH experiments with tou-all probe targeting both tou-RB and the 3′-end of all other tou isoforms in WT and tou-MiMIC larval wing discs.