Figure 1.
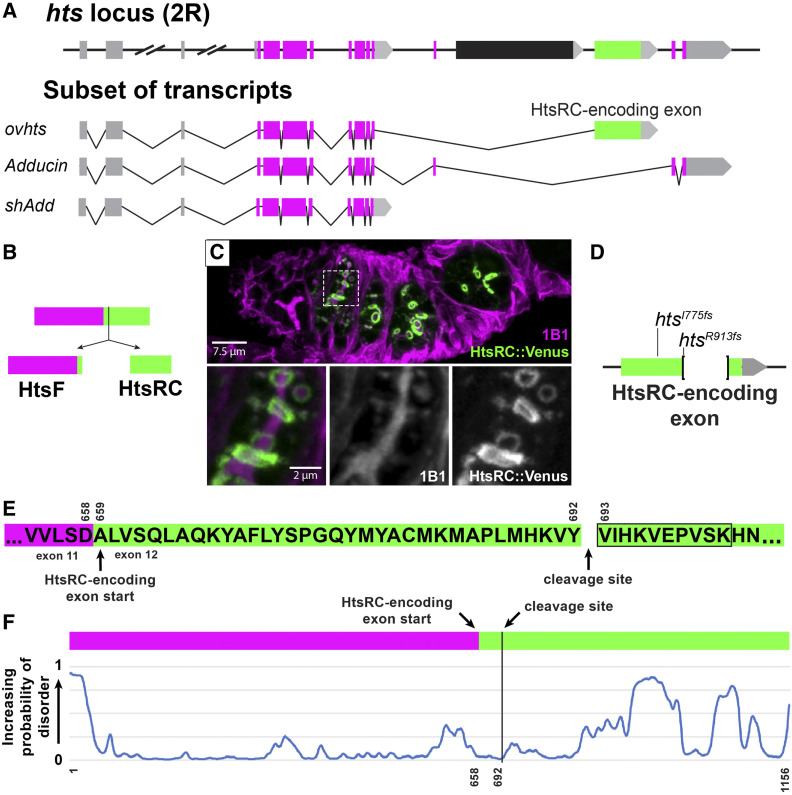
HtsRC is produced from the HtsRC exon after cleavage of the Ovhts polyprotein. (A) Diagram of the hts locus, showing a subset of spliced exons (top) and the structure of three transcripts containing alternatively spliced terminal exons. Untranslated regions and exons are color coded as follows: 5′ UTRs (gray boxes), 3′ UTRs (gray arrows), conserved Adducin exons (magenta boxes), and the HtsRC exon (green box). The black exon corresponds to a testis-specific splice variant not shown. Three hts isoforms with unique terminal exon groups can be detected in the ovary: ovhts, Adducin, and shAdd. Distances are to scale, except where indicated by hash marks. (B) ovhts mRNA produces the Ovhts polyprotein, which is cleaved into HtsF and HtsRC. Exons are color coded as in (A). (C) A germarium expressing transgenic Ovhts::Venus polyprotein. Ovhts tagged at the C-terminus with Venus produces HtsRC::Venus and untagged HtsF following cleavage. HtsF is labeled with a monoclonal antibody (1B1), which also labels Add and shAdd in the fusome. Insets show 1B1 labeling and HtsRC::Venus at the time point where the fusome is dissolving and ring canals first recruit HtsRC. (D) Summary of mutations in the HtsRC exon induced by CRISPR-Cas9 mediated NHEJ. htsI775fs is a 2 bp deletion resulting in a frameshift and 62 novel amino acid residues followed by a stop codon. htsR913fs is a 559 bp deletion followed by a frameshift resulting in 10 novel residues and a stop codon. (E) Amino acid sequence of the Ovhts polyprotein, showing the start of the HtsRC exon and the cleavage site identified by mass spectrometry experiments at residue 693 (residue 35 of the HtsRC exon). Black outline indicates the identified semitryptic peptide. (F) Prediction of intrinsically disordered regions within the Ovhts polyprotein using the SPOT-Disorder-Single application (Hanson et al. 2018). Vertical black line indicates the polyprotein cleavage site described in (E).