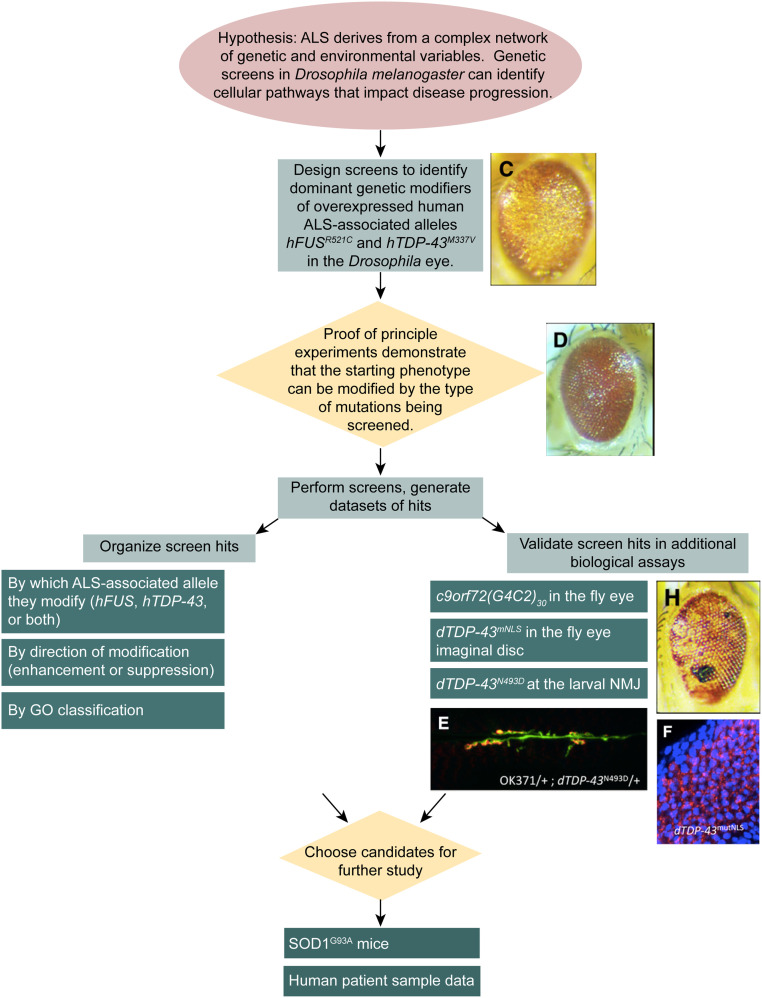
Figure 2.
Workflow diagram illustrating the logic of the screening process. Genetic screens begin with a hypothesis, i.e., that genetic screens in Drosophila can be used to identify novel genes associated with ALS. Because human ALS-associated alleles hFUSR521C or hTDP-43M337V cause a visible degenerative eye phenotype in adult Drosophila, libraries of mutants like the Exelixis collection can be screened for dominant modifiers of this phenotype. Once the screens have been planned, proof-of-principle experiments are conducted to demonstrate that the screen has the potential to be successful, using selected Drosophila mutants of other ALS gene orthologs as positive controls. The screens are then performed, and a list of hits is generated. The list can be organized by which ALS allele is affected by each modifier, whether modifiers are enhancers or suppressors, and according to GO classification for \biological function and/or cellular localization. Screen hits also are validated experimentally by assaying whether they modify other ALS model systems, including c9orf72(G4C2)30-mediated degeneration of the adult Drosophila eye, dTDP-43mNLS aggregation in the larval eye imaginal disc, and dTDP-43N493D perturbation of the larval NMJ. Candidates that show activity in the secondary assays may be chosen for further study, which includes examination in an ALS model mouse and in human ALS patient data.