Fig. 1.
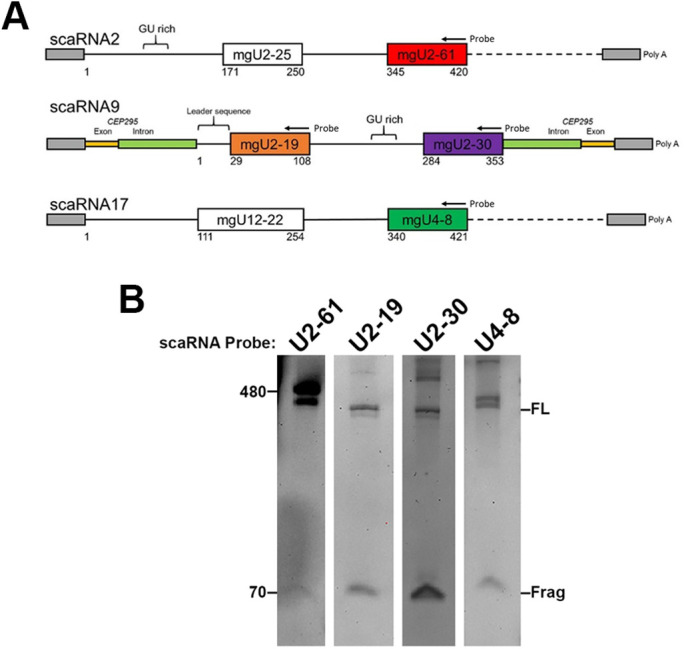
ScaRNA 2, 9 and 17 expression constructs and processing. (A) Schematic of expression constructs used to evaluate in vivo processing of scaRNA 2, 9 and 17. Sequences encoding these scaRNAs were cloned into pcDNA3.1+. Transcription will generate RNAs that include vector sequences at the 5 ′ and 3 ′ ends of the RNA (gray rectangles), including the addition of a poly A tail. The solid black line represents the endogenous full-length (FL) scaRNA detectable by Northern blotting. The dashed line represents sequences downstream of the FL scaRNA that are important for processing. The colored rectangles denote processed endogenous fragments for each scaRNA that are detectable by Northern blotting. In contrast to scaRNA 2 and 17, which are independently transcribed in the genome, scaRNA9 is encoded in the intron of the CEP295 host gene. Our pcDNA3.1+scaRNA9 expression construct contains the entire CEP295 intron that harbors scaRNA9 (green rectangles), along with partial exonic sequences upstream and downstream of this intron (gold rectangles), ensuring proper splicing of the transcribed RNA. The probes used for Northern blotting are indicated. Other notable sequence elements, such as the leader sequence in scaRNA9 and the GU rich regions in scaRNA2 and scaRNA9 (Enwerem et al., 2015; Poole et al., 2016, 2017), are denoted. (B) Detection of ectopically expressed FL and processed fragments by Northern blotting. HeLa cells were transfected with the DNA expression constructions described above, followed by RNA isolation, TBE-Urea gel electrophoresis, Northern blotting and hybridization with probes that bind the indicated region of the scaRNA. The detected signals corresponding to ectopically expressed FL or fragment (frag) for each scaRNA are noted.
