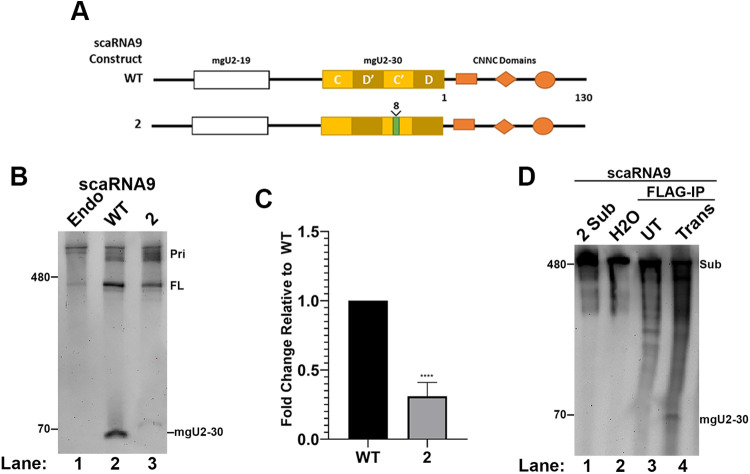
Fig. 5.
Disruption of the scaRNA9 C′ box impedes in vivo biogenesis of the mgU2-30 fragment. (A) Schematic of WT and mutant scaRNA9 constructs. The 8 nt insertion in the C′ box of scaRNA9 mutant 2 is shown. Also indicated are CNNC motifs that are known to impact Drosha/DGCR8 processing. (B) Northern blot detection of the mgU2-30 fragment and FL scaRNA9 in RNA isolated from HeLa cells that ectopically express the WT or mutant #2 scaRNA9 constructs. Primary-scaRNA signal is denoted (Pri). (C) Histogram generated from the quantification of data shown in (B). The mgU2-30 signal was divided by the FL scaRNA9 signal in that lane, and normalized to that obtained for WT. N=4 biological repeats, ****=P<0.0001, error bars represent standard deviation. (D) Northern blot detection of RNA isolated from in vitro processing assays with Drosha/DGCR8 and scaRNA9 mutant #2 as the substrate.