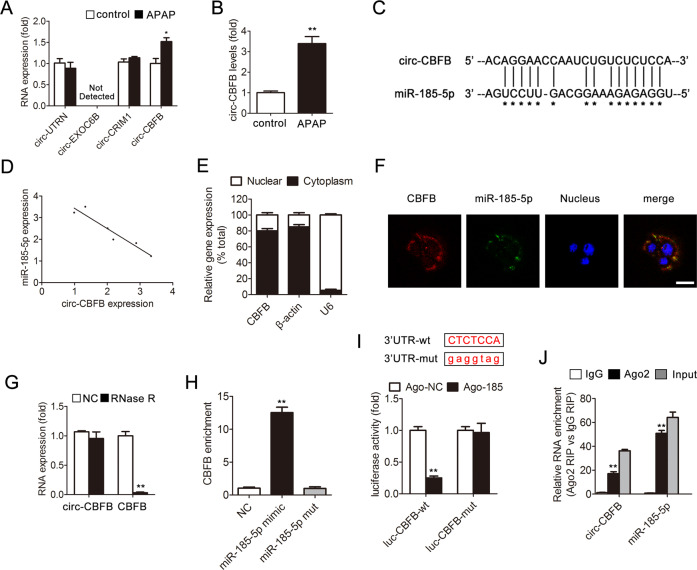
Fig. 6. Characterization of circ-CBFB and its interaction with miR-185-5p.
a Liver circ-UTRN, circ-EXOC6B, circ-CRIM1, circ-CBFB expression, n = 6. b Hepatocyte circ-CBFB expression, n = 6. *p < 0.05, **p < 0.01 vs. the control group. c circ-CBFB has a predicted binding site in miR-185-5p. d The correlation between circ-CBFB and miR-185-5p expression in liver. e Levels of circ-CBFB in the nuclear and cytoplasmic fractions. f Dual-immunofluorescence staining of circ-CBFB and miR-185-5p. Scale bar, 50 μm. g circ-CBFB and CBFB mRNA expression treated with or without RNase R. **p < 0.01 vs. the NC group. h circ-CBFB was pulled down and enriched with a 3′-end biotinylated miR-185-5p mimic. **p < 0.01 vs. the NC group. i AML12 cells were transfected with wild-type or mutant circ-CBFB luciferase constructs and with agomir-NC or miR-185-5p agomir. Luciferase activity was detected 48 h after transfection. **p < 0.01 vs. the ago-NC group. j AGO2 RIP assay of circ-CBFB and miR-185-5p in AML12 cells. circ-CBFB and miR-185-5p expression was detected by qRT-PCR. **p < 0.01 vs. the IgG group.