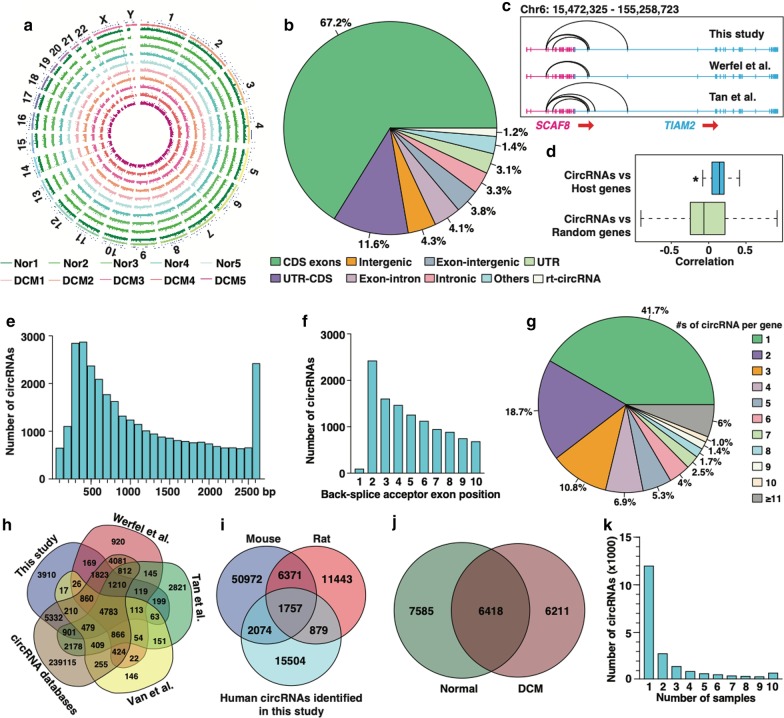
Fig. 1.
General feature of the circRNAs identified in human heart. a The distribution of 20,214 circRNAs identified in human hearts from 5 normal (Nor1-5) and 5 DCM (DCM1-5) patients, was mapped in different chromosomes. The circos plot tracks represent the normalized expression (spliced reads per billion mapping, denoted as SRPBM) of circRNAs in the different samples. b Percentage of genomic origins of the identified circRNAs. c Schematic diagram showing that rt-circRNAs originate from SCAF8 (in red) and TIAM2 (in cyan) genes identified in human heart in this study (upper) and in studies by Werfel et al. (middle) [29] and Tan et al. [27] (bottom). Arrows point to the transcriptional directions of SCAF8 and TIAM2 genes. d Correlation between the expression of circRNAs and their host genes or randomly chosen linear genes. *p < 0.05. e The size distribution of identified circRNAs. f Distribution of circRNAs with respect to most upstream circularized exon. g Distribution of numbers of circRNAs produced per gene. h Four-set venn diagram showing the over-lapping circRNAs identified in human heart in this study and in studies by Tan et al., Werfel et al. and Van et al., as well as 263,738 non-redundant circRNAs obtained by merging human circRNAs in 4 circRNA databases including circBase (n = 91,986), circBank (n = 140,331), circRNADB (n = 32,883), and CIRCpedia V2 (n = 183,943). i Venn diagram showing the over-lapping circRNAs identified in human heart in this study and reported mouse and rat circRNAs. The 61,174 non-redundant mouse circRNAs were obtained by merging circRNAs reported by study of Werfel et al. (n = 9474), Jakobi et al. (n = 561), and Tan et al. (n = 2632), as well as 2 circRNA databases including circBase (n = 1756) and CIRCpedia V2 (n = 54,274). The 20,450 non-redundant rat circRNAs were obtained by merging circRNAs reported by study of Werfel et al. (n = 12,256) and CIRCpedia V2 (n = 17,083). j Venn diagram showing the number of circRNAs unique or common between normal and DCM human samples. k Number of human heart samples in which circRNAs with different abundance were identified