Abstract
In response to the current coronavirus disease 2019 (COVID-19) pandemic, it is crucial to understand the origin, transmission, and evolution of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which relies on close surveillance of genomic diversity in clinical samples. Although the mutation at the population level had been extensively investigated, how the mutations evolve at the individual level is largely unknown. Eighteen time-series fecal samples were collected from nine patients with COVID-19 during the convalescent phase. The nucleic acids of SARS-CoV-2 were enriched by the hybrid capture method. First, we demonstrated the outstanding performance of the hybrid capture method in detecting intra-host variants. We identified 229 intra-host variants at 182 sites in 18 fecal samples. Among them, nineteen variants presented frequency changes > 0.3 within 1–5 days, reflecting highly dynamic intra-host viral populations. Moreover, the evolution of the viral genome demonstrated that the virus was probably viable in the gastrointestinal tract during the convalescent period. Meanwhile, we also found that the same mutation showed a distinct pattern of frequency changes in different individuals, indicating a strong random drift. In summary, dramatic changes of the SARS-CoV-2 genome were detected in fecal samples during the convalescent period; whether the viral load in feces is sufficient to establish an infection warranted further investigation.
Keywords: SARS-CoV-2, Intra-host variant, Dynamics, Mutation, Hybrid capture
1. Introduction
The ongoing coronavirus disease 2019 (COVID-19) pandemic has brought a severe threat to public health and the global economy. The causative pathogen, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is likely to be of zoonotic origin. Genomes of RNA viruses mutate fast and undergo rapid evolution, which could ultimately affect their virulence, infectivity, and transmissibility. Monitoring variations in the SARS-CoV-2 genome at the population level is vital for tracing the outbreak origin, tracking transmission chains, and understanding viral evolution (Forster et al., 2020; Lu et al., 2020; van Dorp et al., 2020). Meanwhile, the study of intra-host single-nucleotide variation (iSNV), which represents the intermediate stage between the origin and the fixation of the mutation at the individual level, is essential for understanding how the virus evolves in the human body under immune pressure (Domingo et al., 2012). These studies require complete, in-depth, and unbiased profiling of SARS-CoV-2 genomes from a large number of patients.
However, there are still challenges in obtaining a high-quality SARS-CoV-2 genome directly from clinical samples, especially for those with low viral loads. For samples with Ct > 30, the proportion of genome recovered was lower than 90%, irrespective of the amplification and sequencing approach used (Lu et al., 2020). Current strategies for targeted enrichment of the SARS-CoV-2 genome include hybrid capture and multiplex polymerase chain reaction (PCR) amplification; the former method was proposed to be more reliable in generating unbiased coverage across the genome and identifying minor alleles (Xiao et al., 2020b). As the effectiveness of the hybrid capture method is influenced by multiple factors, such as probe design, rounds of hybridization, and viral load, the fidelity of hybrid capture warrants further evaluation in the context of iSNV investigation.
Recent studies have revealed that the gastrointestinal (GI) tract is an important factor in the pathogenesis and transmission of COVID-19. GI infection of SARS-CoV-2 has been confirmed by the isolation of viable virus from the fecal specimen (Xiao et al., 2020a; Zhou et al., 2020), and diarrhea is observed in > 20% of hospitalized patients with COVID-19 (Wan et al., 2020). Moreover, the shedding of SARS-CoV-2 from the GI tract lasted much longer than that from the respiratory tract in children (Xu et al., 2020), raising the possibility of fecal-oral transmission. However, neither the genomic diversity of the virus nor its longitudinal dynamics in the GI tract is known.
To disentangle how SARS-CoV-2 evolves and adapts in the GI tract during the convalescence period, 18 longitudinal fecal specimens from nine children with COVID-19 were collected. Nucleic acids of SARS-CoV-2 were enriched using a hybrid capture method. Seventeen near-complete genomes (> 99% genome covered) of SARS-CoV-2 were obtained. Among them, the majority rule consensus sequences at four sites were changed in 1–5 days, and another 15 sites showed allele frequency changes higher than 0.3, indicating a rapid genomic change in the fecal samples.
2. Results
2.1. Hybrid capture effectively enhanced SARS-CoV-2 reads and genome coverage
Eighteen fecal samples were collected from nine children with COVID-19, and two samples were collected from each child within a time interval of 1–5 days (T1 and T2). Ages of the children ranged from three months to 13 years. Their symptoms were either mild or moderate, and two of them had diarrhea. All samples were collected during the recovery stage (see Table S1 for more details). We performed metatranscriptomic sequencing (Raw) and hybrid capture-based sequencing (with one round of hybridization, E1) on all samples. Meanwhile, to investigate the efficiency of hybridization, nine samples were processed with an extra round of hybridization (E2). Two fecal samples from two healthy children were used as negative controls (NCs). More than 25 million reads were generated for each sample, except that no less than 12 million reads were generated for each NC sample. For COVID-19 samples, Ct values of RT-qPCR targeting SARS-CoV-2 varied from 28 to lower than the detection limit, with a median of 33.26.
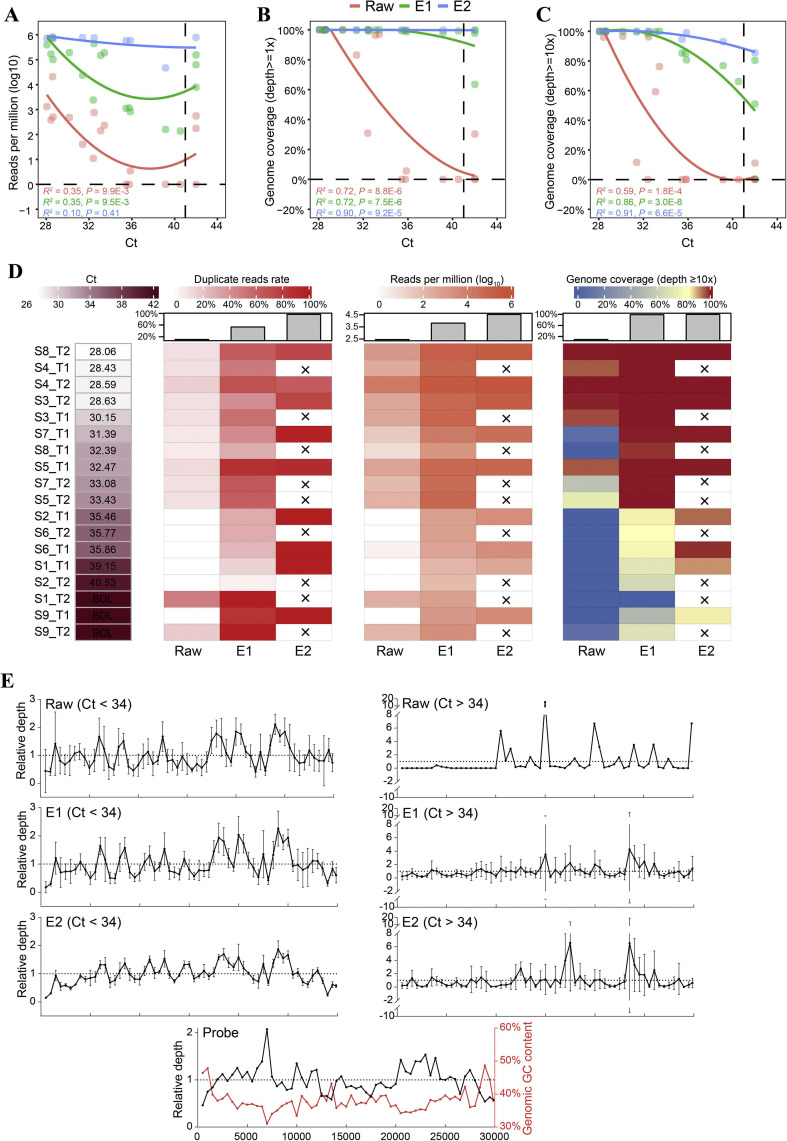
A median number of 157 (range = 0–20,816) reads per million (RPM) reads were mapped to the SARS-CoV-2 reference genome in Raw data. As expected, the number of SARS-CoV-2 reads was negatively correlated with the Ct value (Spearman' ρ = −0.59, P < 0.01, Fig. 1 A). One round of hybrid capture greatly improved the number of SARS-CoV-2 reads in all samples (RPM range = 136–763,746, median = 82,137, Wilcoxon matched-pairs signed rank test, P < 0.0001). An extra round of hybridization enabled further improvement (RPM = 47,072-810,801, median = 718,025, P < 0.01), which was 19,726 and 33 times higher than those in Raw and E1 data, respectively (Fig. 1A). Consequently, the hybrid capture method significantly increased SARS-CoV-2 genome coverage (Fig. 1B and C; Table 1 ). In addition, we detected 24 and 12 SARS-CoV-2 RPM in two NC samples with one round of hybrid capture, indicating that the signal of SARS-CoV-2 observed in the patients with COVID-19 was much stronger than that in the NC.
Fig. 1.
SARS-CoV-2 read counts and genome coverage obtained using direct metatranscriptomic sequencing and hybrid capture-based sequencing. A: The number of SARS-CoV-2 reads in the unit of reads per million (RPM). B: Genome coverage with depth ≥1. C: Genome coverage with and depth ≥ 10. In A–C, local polynomial regression line, R, and p values of Spearman correlations are shown; the Ct value that was below the detection limit was replaced with 42 for better visualization. D: Heatmap of Ct, duplicate reads rate, RPM, and genome coverage after deduplication. The median number for each group is shown with a bar plot, and crosses indicate that the samples were not included in E2 data. BDL, below the detection limit. E: Depth distribution of samples and probes along the SARS-CoV-2 genome. The relative depth was calculated by normalizing depth (bin size = 500 bases) to the average depth of each sample or the probes. Samples with Ct < 34 and Ct > 34 are shown separately; only samples with average depth > 1 are shown. The red curve indicates GC content along the reference genome. SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Table 1.
Coverage of the SARS-CoV-2 genome obtained using direct metatranscriptomic sequencing and hybrid capture-based sequencing.
| Group/coverage (range, median, %) |
Before deduplication |
After deduplication |
||||
|---|---|---|---|---|---|---|
| Depth ≥ 1 | Depth ≥ 10 | Depth ≥ 50 | Depth ≥ 1 | Depth ≥ 10 | Depth ≥ 50 | |
| Raw | 0.0–100.0, 57.0 | 0.0–100.0, 11.4 | 0–99.9, 0.9 | 0.0–100.0, 57.0 | 0.0–100.0, 9.2 | 0–99.9, 0.4 |
| E1 | 63.5–100.0, 99.9 | 0.3–100.0, 99.1 | 0.2–100.0, 94.6 | 63.5–100.0, 99.9 | 0.3–100.0, 98.9 | 0.1–100.0, 92.4 |
| E2 | 99.7–100.0, 99.9 | 85.5–100.0, 99.9 | 22.8–100.0, 99.7 | 99.7–100.0, 99.9 | 85.1–100.0, 99.7 | 18.1–100.0, 99.7 |
SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
An excess number of PCR cycles were implemented to ensure enough input for sequencing after the hybridization owing to the loss of nontarget nucleic acids. Accordingly, we noted that the proportion of duplicated reads increased from 10.9% in the Raw to 53.1% after the first hybridization and 95.5% after the second hybridization (Fig. 1D). Despite the higher proportion of duplicate reads, the valid number of SARS-CoV-2 reads and genome coverage after deduplication still significantly increased after hybrid capture (E1 vs. Raw: median fold change = 246.2, E2 vs. E1: 2.7; Fig. 1D; Table 1). Through our calculation, for samples with Ct < 29, direct metatranscriptomic sequencing (with 6G data) is sufficient to obtain a complete viral genome with a sequencing depth of at least 10-fold. One and two rounds of hybrid capture are needed for samples with a Ct of 29–34 and Ct of 34–39 to cover the complete genome, respectively. By contrast, it is challenging to obtain adequate genome coverage for samples with Ct > 39 even when using two rounds of hybrid capture.
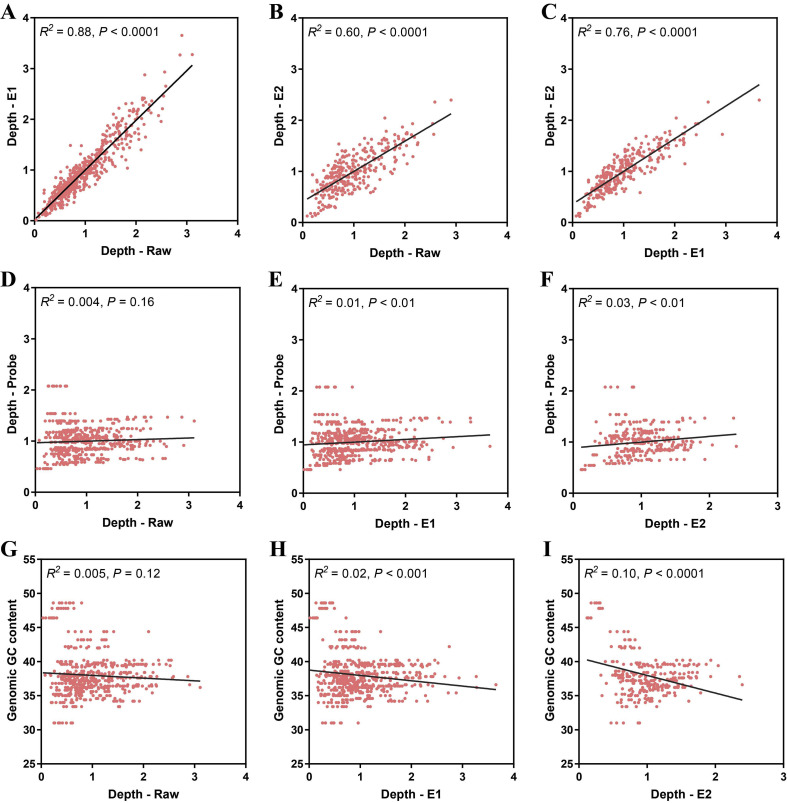
Besides the sequencing coverage, the sequencing depth distribution is another critical metric for downstream analysis; thus, we examined whether the depth distribution is well maintained after hybrid capture. For samples with Ct < 34, similar depth distributions were observed between the Raw data and capture enriched data (linear regression, Raw vs. E1, R 2 = 0.88, Raw vs. E2, R 2 = 0.60, E1 vs. E2, R 2 = 0.76, P < 0.0001; Figs. 1E, S1A–C). Of note, the depth distribution was only slightly influenced by the probe density and genomic GC content, as the correlation was relative low between the depth and probe density (linear regression, E1, R 2 = 0.01; E2, R 2 = 0.03; P < 0.01; Fig. S1D–F) and between the depth and GC content (E1, R 2 = 0.02, P < 0.001; E2, R 2 = 0.10, P < 0.0001; Fig. S1G–I). The standard deviation of depths among samples decreased from Raw to E1 (median = 0.40 and 0.29, Wilcoxon matched-pairs signed rank test, P < 0.001) and further to E2 (median = 0.14, Wilcoxon matched-pairs signed rank test, P < 0.0001), demonstrating a consistent recovery of the viral genome with hybrid capture. For samples with Ct > 34, hybrid capture resulted in unexpected depth peaks (17,000–17,500 and 23,500–24,000) that could not be explained by the probe density or the GC content.
2.2. Hybrid capture enabled reliable inter-host and intra-host variant profiling
By comparing the consensus sequences (major allele frequency > 0.7) of E1/E2 with those of Raw, we identified 41 inconsistent sites (with depth > 5) (Table S2). However, all discrepancies involved in intra-host variants (the frequency of the major allele in at least one data set < 0.7), and only four of them involved in major allele switches with frequency changes varying from 0.17 to 0.54 (three sites with depth < 10), suggesting high reliability of the genome obtained with hybrid capture.
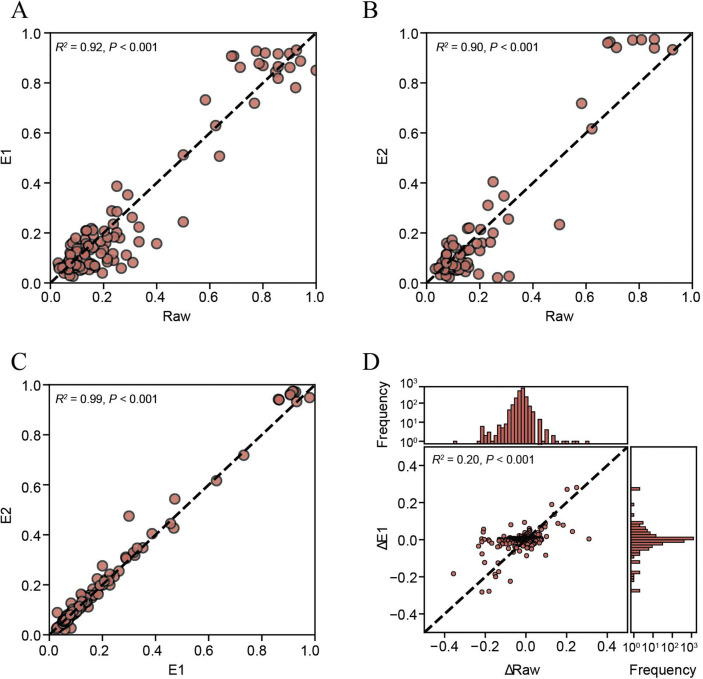
We further compared the alternative allele frequencies (AAFs) among Raw, E1, and E2. Although the number of mutations detected in Raw was much less than that in E1 (11 vs. 216), which were caused by the limited genome coverage in Raw, there were decent linear correlations among AAFs of Raw, E1, and E2 (Raw vs. E1, R 2 = 0.92, Raw vs. E2, R 2 = 0.90, E1 vs. E2, R 2 = 0.99, P < 0.001; Fig. 2 A–C). Moreover, the change of AAFs from T1 to T2 was also significantly correlated between Raw and E1 (R 2 = 0.20, P < 0.001; Fig. 2D). Notably, the frequency change calculated from the Raw data showed a larger variance than E1 (Fig. 2D), likely reflecting more significant stochastic fluctuations related to the lower sequencing depth. These results demonstrated that hybrid capture-based sequencing is ideal for genomic diversity analysis and also enable an accurate longitudinal analysis of iSNVs.
Fig. 2.
Evaluation of hybrid capture in mutant allele frequency profiling. A–C: Consistency among alternative allele frequencies (AAFs) in Raw, E1, and E2 data. Mutations were identified as described in the Materials and methods section; if a mutation was only identified in one of the comparison pairs, the AAF of the other one was used as long as its depth ≥ 10. D: Consistency between AAF changes from T1 to T2 in Raw and E1 data. Sites with depth ≥ 10, minor allele frequency ≥ 0.01, and minor allele supporting reads ≥ 2 were used. R2 and p values of linear regressions are shown.
2.3. iSNV characteristics and longitudinal dynamics in fecal samples
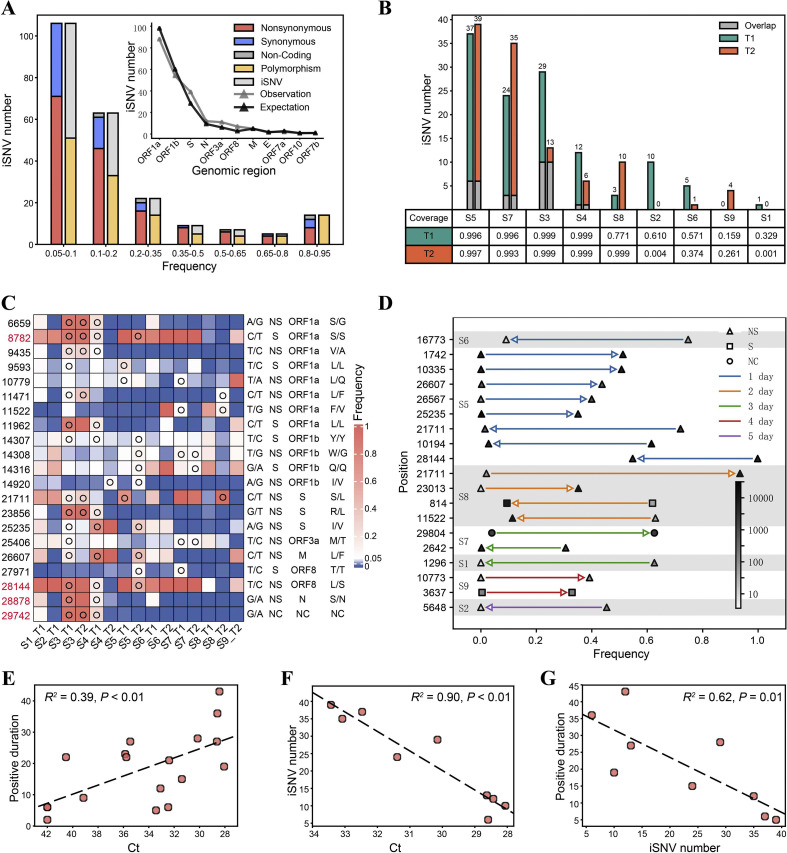
Taking advantage of the higher sequencing depth and high fidelity of the SARS-CoV-2 genome obtained using the hybrid capture method, E1 and E2 data were pooled together for iSNV analysis. A total number of 229 iSNVs were identified at 182 sites (criteria are described in Materials and methods), including 159 nonsynonymous, 60 synonymous, 3 noncoding, and 7 stop-gain mutations. The overall Ka:Ks ratio was 0.54, which was significantly different from one (Fisher's exact test, P < 0.001), and thus suggested a purifying selection. The number of iSNVs was inversely correlated with AAF, except for iSNVs with an AAF of 0.8–0.95 (Fig. 3 A), which probably reflected the back mutation from the mutant allele to the ancestral allele. Besides, the number of iSNVs in different genes is proportional to the length of the corresponding regions (Fig. 3A), suggesting no visible mutation hot spot on the SARS-CoV-2 genome. We noted that 54% (124 of 229) of the mutations were polymorphic in the population (by comparing with 20,789 mutations retrieved from https://bigd.big.ac.cn/ncov, September 12, 2020) (Zhao et al., 2020), and the AAF of iSNVs that were polymorphic in the population was higher than that of other iSNVs (Chi-square test, P = 0.01; Fig. 3A), which is in accordance with the principle that the chance of getting fixed and transmitted is proportional to the frequency of the mutation in an individual. Notably, all 14 mutations with an AAF of 0.8–0.95 were polymorphic in the population.
Fig. 3.
iSNV profiles and longitudinal dynamics in fecal samples. A: Alternative allele frequency (AAF) distribution of all iSNVs. The inserted plot shows the number of observed iSNVs in different genes; the expected number of iSNVs was calculated based on the length of each region. B: The number of iSNVs identified in each sample. Genomic coverages with depth > 50 are shown below the plot. C: AAF at iSNV positions shared by multiple individuals. The nucleotide at the position (reference allele/alternative allele), mutation type, genomic region, and amino acid change are shown on the right of the heatmap. Open circles indicate identified iSNVs. The four positions associated with recurrent mutations (van Dorp et al., 2020) are highlighted in red. D: iSNVs with frequency change > 0.30 from T1 to T2. Only changes with adjusted P < 0.05 (Fisher's exact test) are shown. Arrows indicate the direction of changes from T1 to T2, the colors of lines indicate time intervals between T1 and T2, and the colors of triangles/squares/dots indicate the number of nonduplicated reads at each site. E–G: Correlation between the Ct value, iSNV number, and the duration of detection of SARS-CoV-2 RNA in feces since T1/T2. R2 and p values of linear regressions are shown. In F and G, only samples having full genomic coverage with depth ≥ 50 were included (n = 9). NS, nonsynonymous, S, synonymous, NC, noncoding; iSNV, intra-host single-nucleotide variation.
The number of iSNVs markedly varied among different individuals, ranging from 6 to 39 in samples with the full genome covered by at least 50-fold (Fig. 3B). Twenty-one iSNVs were shared among multiple individuals, which is more than expected by chance (Exact Poisson test, P < 0.001; Fig. 3C). Moreover, four of the shared iSNVs were located at mutation hot spots (van Dorp et al., 2020). Considering that only 198 such mutation hot spots were identified, shared iSNVs were significantly enriched at positions with higher mutation rates (Exact Poisson test, P < 0.001). C8782T and T28144C are widespread variants that defined a subgroup of SARS-CoV-2 (Tang et al., 2020; Wang et al., 2020b); they were both heterogeneous in multiple samples in our data, suggesting that either the position is prone to mutate or there existed simultaneous transmission of multiple strains in the population when the samples were collected. Mutations at most positions only involved transitions between two nucleotides, except that more than two nucleotides were observed at four positions, namely, 14,307, 14,308, 11,147, and 11,148 (Fig. 3C; Table S3).
The dynamics of all iSNVs were then examined, and only 8.3–34.5% of iSNVs were observed at both T1 and T2 (Fig. 3B). Meanwhile, 19 iSNVs showed frequency changes higher than 0.30 within 1–5 days (Fig. 3D), 16 of which were nonsynonymous mutations. Nine of these significant shifts occurred within a day, and four occurred within two days, implying intense selection pressure or a strong genetic drift. Notably, the most dramatically shifted iSNV C21771T, which caused a nonsynonymous mutation from Ser to Leu, was observed in two individuals but showed opposite tendencies, whereby the frequency of the mutant allele increased by 0.91 in one day in S8 but decreased by 0.71 in two days in S5. Although we favored a random genetic drift hypothesis, considering that the viral genome differed at more than 20 positions between S8 and S5, the possibility of distinct selection pressures under different genomic backgrounds cannot be ruled out. Besides, the number of rapidly changing positions varied among different individuals, which may reflect different immune pressures (Fig. 3D). We also noted that some mutations in the same individual had similar frequencies and showed concurrent changes (S5 and S8), suggesting they were located on the same haplotype.
We further investigated the correlation between virological features and clinical features. First, the load of SARS-CoV-2 was found to be positively correlated with the duration of detection of SARS-CoV-2 RNA in feces since T1/T2 (linear regression, R 2 = 0.39, P < 0.01; Fig. 3E). Second, the number of iSNV was in a negative correlation with the SARS-CoV-2 load (R 2 = 0.90, P < 0.01; Fig. 3F), as well as with the duration of detection of SARS-CoV-2 RNA (R 2 = 0.62, P = 0.01; Fig. 3G). Such a negative correlation between the viral load and genomic diversity was also observed in patients with COVID-19 and cancer (Siqueira et al., 2020). We speculate that the lower viral load reflects stronger immune pressure, which may promote adaptive evolution and result in higher intra-host diversity. This was reported in the chronic infection of HIV and HCV in humans (Farci et al., 2000; Yu et al., 2018). However, because the virus was quickly cleared in patients with greater intra-host diversity, the underlying mechanism seems different and warrants further study.
3. Discussion
The limited viral load in clinical samples has long been an obstacle to both pathogen detection and genomic studies, especially considering that the abundance of viruses could decrease when the disease progresses. Here, we have demonstrated that the hybrid capture method was capable of recovering the unbiased SARS-CoV-2 genome from RT-qPCR-negative and metatranscriptomic sequencing-negative samples. Furthermore, it also enables reliable analysis of inter-host and intra-host variants, making it a promising strategy for genomic studies on SARS-CoV-2.
The identification of iSNVs in fecal samples, as well as in throat swabs and bronchoalveolar lavage fluid, suggests ongoing evolution of SARS-CoV-2 in the human body (Rose et al., 2020; Sashittal et al., 2020; Shen et al., 2020; Wang et al., 2020a). However, the possibility of infection of multiple strains cannot be entirely ruled out. With the time-series samples, we have proved that novel mutations could occur within one day, and the frequency of the mutation changed notably fast in humans. Thus, we speculate that iSNVs are more likely to represent spontaneous mutations rather than infection of multiple strains. A recent study proposed that SARS-CoV-2 in samples from the GI tract had a relatively higher genomic diversity than that from the respiratory tract (Wang et al., 2020a), suggesting that the mutation rate may vary among different tissues. Thus, the fast evolution of the viral genome in fecal samples observed in our study may not be applicable to samples from other organs.
All samples in this study were collected from pediatric patients in rehabilitation with no clinical symptoms. There had been more than two weeks since symptom onset for most patients except S5 (with a median of 24 days). Fecal shedding of SARS-CoV-2 had been observed in multiple studies and was proposed to be longer than respiratory samples (Santos et al., 2020; Xu et al., 2020). However, whether RNA shedding from stools is infectious during the convalescent phase is unclear (Amirian, 2020). The evolution of the viral genome in feces observed in our study suggested that the virus was viable, and probably, the virus actively replicated in the GI tract. Thus, we speculate that the fecal transmission of SARS-CoV-2 is possible, and further study is warranted to investigate whether the viral load in feces is sufficient to establish an infection.
Although a general purifying selection on the viral genome had been shown in our data and also by previous studies, we did not observe any signature of selection on specific positions. Wang et al. (2020a) found increased frequencies of two mutations (C21711T and G11083T) in two samples and suspected an adaptive selection on these mutations. Interestingly, C21711T was also observed in our data and showed the greatest frequency changes. However, instead of simultaneous increases, the direction of the frequency change was opposite in two samples, indicating a robust random drift effect. Of note, the viral genome of the two samples showed significant divergence (> 20 substitutions), and how the genomic background interacts with a single mutation needs further investigation.
C8782T and T28144C had been reported to be in linkage disequilibrium and used to define an ancestral subgroup of SARS-CoV-2 (Tang et al., 2020). The authors interpreted the heteroplasmy status at these two sites as evidence for multiple-strain infection. However, our data indicated that these two positions were prone to mutate, and their frequencies were subject to change. van Dorp et al. (2020) also identified these two positions as hot spots for recurrent mutations. Thus, the grouping of SARS-CoV-2 genomes based on these two mutations should be with great caution.
The mutation and their frequency change may also affect the response to drugs. Position 10,194 locates in the region encoding 3-chymotrypsin-like cysteine protease (3CLpro), which is essential for the replication of coronavirus, and this region is also predicted to be the target for a few drugs including lopinavir (Das et al., 2020; Jin et al., 2020). At the first time point in patient S5, there was an equivalent number of reads presenting A and T at this position (a mutation from A to T results in an amino acid change from Glu to Val). At the second time point, which was one day later, the mutant allele T almost disappeared. Although it is unclear whether the mutation could affect the binding of antiviral drugs, the risk should not be overlooked.
There are limitations in this study. SARS-CoV-2 could potentially infect cells in multiple organs (Prasad and Prasad, 2020; Xiao et al., 2020a), and the viral RNA in feces may be derived from the GI tract or other organs. The evolution of the virus in different organs or different locations in the same organ is independent, and to what extent they vary is still unknown (Domingo et al., 2012). As we only used an aliquot of 200 mg of homogenized feces as the starting material, the diversity of the viral genome may be underrepresented and biased. Biological replicates should be included to evaluate the sampling bias in future studies.
In summary, our study highlighted the need for extensive studies on the intra-host variant dynamics in different tissues, with large cohorts spanning a wide range of ages, disease severity, and geographic regions.
4. Materials and methods
4.1. Sample collection and ethics
Eighteen fecal samples were collected from nine hospitalized children with quantitative reverse transcription PCR (RT-qPCR)-confirmed SARS-CoV-2 infection. Two fecal samples were collected from each child within a time interval of 1–5 days (T1 and T2) in the recovery stage. The patients were characterized by mild symptoms and long duration of SARS-CoV-2 shedding in their feces (see Table S1 for demographic and clinical information). All samples were inactivated at 56°C for 30 min and stored at −80°C before processing.
This study was approved by the ethics review committee of Guangzhou Women and Children's Medical Center. Written informed consents were obtained from the legal guardians of all children.
4.2. Metatranscriptomic and hybrid capture-based sequencing
Each fecal sample (~200 mg) was suspended in 1 mL of Phosphate Buffered Saline (PBS) and centrifuged at 8000 g for 5 min to obtain the supernatant. RNA was extracted from 200 μL of the supernatant (AllPrep® PowerViral® DNA/RNA Kit; Qiagen, Hilden, Germany), concentrated (RNA Clean and Concentrator™-5 with DNase I; Zymo Research, Irvine, CA), and used for library preparation (Trio RNA-Seq; Nugen, Redwood City, CA). An aliquot of 750-ng library from each sample was used for hybrid capture-based enrichment of SARS-CoV-2 with one or two rounds of hybridization (1210 ssRNA probes, TargetSeq® One nCov Kit; iGeneTech, Beijing, China). Sequencing was performed on the Illumina HiSeq X Ten platform. The load of SARS-CoV-2 was quantified using the RT-qPCR targeting ORF1ab gene (Real-Time Fluorescent RT-PCR Kit for Detecting, 2019-nCoV; BGI, Wuhan, China).
4.3. Sequencing data analysis
Quality control of the sequencing reads including adapter trimming, low-quality reads removal, and short reads removal was performed using fastp version 0.20.0 (Chen et al., 2018) (-l 70, -x, -cut-tail, -cut_tail_mean_quality 20). Clean reads were mapped to the SARS-CoV-2 reference genome Wuhan-Hu-1 (GenBank MN908947.3) using BWA mem version 0.7.12 (Li, 2013), followed by duplicate reads removal using Picard version 2.18.22 (http://broadinstitute.github.io/picard). Mpileup files were generated using samtools version 1.8 (Li et al., 2009). Intra-host variants were identified using VarScan version 2.3.9 and an in-house script (Koboldt et al., 2012). Criteria for variants included the following: (1) sequencing depth ≥ 50, (2) minor allele frequency ≥ 5%, (3) minor allele frequency ≥ 2% on each strand, (4) minor allele counts ≥ 10 on each strand, (5) strand bias of the minor allele < 10-fold, (6) minor allele was supported by the inner part of the read (excluding 10 base pairs on each end), and (7) minor allele was supported by ≥ 10 reads that were classified as Betacoronavirus by Kraken version 2.0.8-beta (Wood et al., 2019) on each strand.
The ratio of the number of nonsynonymous substitutions per nonsynonymous site (Ka) to the number of synonymous substitutions per synonymous site (Ks) was calculated using KaKs_Calculator 2.0 (Wang et al., 2010) by the MS method.
4.4. Data availability
Viral reads were deposited in the Genome Warehouse in the National Genomics Data Center (National Genomics Data Center Members and Partners, 2020) under project PRJCA2828, which are publicly accessible at https://bigd.big.ac.cn/gsa.
CRediT authorship contribution statement
Yi Xu: Conceptualization, Resources, Supervision. Lu Kang: Methodology, Data curation, Formal analysis. Zijie Shen: Methodology, Data curation, Formal analysis. Xufang Li: Methodology, Resources. Weili Wu: Methodology, Investigation. Wentai Ma: Methodology, Data curation. Chunxiao Fang: Methodology, Resources. Fengxia Yang: Methodology, Resources. Xuan Jiang: Methodology. Sitang Gong: Supervision, Resources. Li Zhang: Methodology, Data curation, Formal analysis, Supervision, Writing - original draft preparation, Writing-Reviewing and Editing. Mingkun Li: Conceptualization, Supervision, Writing- Reviewing and Editing.
Acknowledgments
We thank Dr. Yongbiao Xue and colleagues from the National Genomics Data Center for helpful discussion and computational resource support. We thank Dr. Hong Tang and Dr. Dongping Liu from Institut Pasteur of Shanghai, Chinese Academy of Sciences, for project coordination. This work was supported by grants from National Key R&D Program of China (2020YFC0848900), the Strategic Priority CAS Project (XDB38000000), Chinese Academy of Sciences and the National Major Science and Technology Project for Control and Prevention of Major Infectious Diseases in China (2018ZX10305409, 2018ZX10301401, 2018ZX10732401).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jgg.2020.10.002.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
References
- Amirian E.S. Potential fecal transmission of SARS-CoV-2: current evidence and implications for public health. Int. J. Infect. Dis. 2020;95:363–370. doi: 10.1016/j.ijid.2020.04.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Zhou Y., Chen Y., Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S., Sarmah S., Lyndem S., Singha Roy A. An investigation into the identification of potential inhibitors of SARS-CoV-2 main protease using molecular docking study. J. Biomol. Struct. Dyn. 2020:1–11. doi: 10.1080/07391102.2020.1763201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo E., Sheldon J., Perales C. Viral quasispecies evolution. Microbiol. Mol. Biol. Rev. 2012;76:159–216. doi: 10.1128/MMBR.05023-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farci P., Shimoda A., Coiana A., Diaz G., Peddis G., Melpolder J.C., Strazzera A., Chien D.Y., Munoz S.J., Balestrieri A., Purcell R.H., Alter H.J. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science. 2000;288:339–344. doi: 10.1126/science.288.5464.339. [DOI] [PubMed] [Google Scholar]
- Forster P., Forster L., Renfrew C., Forster M. Phylogenetic network analysis of SARS-CoV-2 genomes. Proc. Natl. Acad. Sci. U. S. A. 2020;117:9241–9243. doi: 10.1073/pnas.2004999117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- Koboldt D.C., Zhang Q., Larson D.E., Shen D., McLellan M.D., Lin L., Miller C.A., Mardis E.R., Ding L., Wilson R.K. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 3, 13033997. 2013 [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;1000:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., du Plessis L., Liu Z., Hill V., Kang M., Lin H., Sun J., François S., Kraemer M.U.G., Faria N.R., McCrone J.T., Peng J., Xiong Q., Yuan R., Zeng L., Zhou P., Liang C., Yi L., Liu J., Xiao J., Hu J., Liu T., Ma W., Li W., Su J., Zheng H., Peng B., Fang S., Su W., Li K., Sun R., Bai R., Tang X., Liang M., Quick J., Song T., Rambaut A., Loman N., Raghwani J., Pybus O.G., Ke C. Genomic epidemiology of SARS-CoV-2 in Guangdong Province, China. Cell. 2020;181:997–1003. doi: 10.1016/j.cell.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Genomics Data Center Members and Partners Database resources of the national genomics data center in 2020. Nucleic Acids Res. 2020;48:D24–D33. doi: 10.1093/nar/gkz913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad A., Prasad M. Single virus targeting multiple organs: what we know and where we are heading? Front. Med. 2020;7:370. doi: 10.3389/fmed.2020.00370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose R., Nolan D.J., Moot S., Feehan A., Cross S., Garcia-Diaz J., Lamers S.L. Intra-host site-specific polymorphisms of SARS-CoV-2 is consistent across multiple samples and methodologies. medRxiv. 2020 2020.04.24.20078691. [Google Scholar]
- Santos V.S., Gurgel R.Q., Cuevas L.E., Martins-Filho P.R. Prolonged fecal shedding of SARS-CoV-2 in pediatric patients. A quantitative evidence synthesis. J. Pediatr. Gastroenterol. Nutr. 2020;71(2):150–152. doi: 10.1097/MPG.0000000000002798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sashittal P., Luo Y., Peng J., El-Kebir M. Characterization of SARS-CoV-2 viral diversity within and across hosts. bioRxiv. 2020 2020.05.07.083410. [Google Scholar]
- Shen Z., Xiao Y., Kang L., Ma W., Shi L., Zhang L., Zhou Z., Yang J., Zhong J., Yang D., Guo L., Zhang G., Li H., Xu Y., Chen M., Gao Z., Wang J., Ren L., Li M. Genomic diversity of SARS-CoV-2 in coronavirus disease 2019 patients. Clin. Infect. Dis. 2020;71:713–720. doi: 10.1093/cid/ciaa203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siqueira J.D., Goes L.R., Alves B.M., de Carvalho P.S., Cicala C., Arthos J., Viola J.P.B., de Melo A.C., Soares M.A. SARS-CoV-2 genomic and quasispecies analyses in cancer patients reveal relaxed intrahost virus evolution. bioRxiv. 2020 doi: 10.1093/ve/veab013. 2020.08.26.267831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Wu C., Li X., Song Y., Yao X., Wu X., Duan Y., Zhang H., Wang Y., Qian Z., Cui J., Lu J. On the origin and continuing evolution of SARS-CoV-2. Natl. Sci. Rev. 2020;7:1012–1023. doi: 10.1093/nsr/nwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dorp L., Acman M., Richard D., Shaw L.P., Ford C.E., Ormond L., Owen C.J., Pang J., Tan C.C.S., Boshier F.A.T., Ortiz A.T., Balloux F. Emergence of genomic diversity and recurrent mutations in SARS-CoV-2. Infect. Genet. Evol. 2020;83:104351. doi: 10.1016/j.meegid.2020.104351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Li J., Shen L., Zou Y., Hou L., Zhu L., Faden H.S., Tang Z., Shi M., Jiao N., Li Y., Cheng S., Huang Y., Wu D., Xu Z., Pan L., Zhu J., Yan G., Zhu R., Lan P. Enteric involvement in hospitalised patients with COVID-19 outside Wuhan. lancet. Gastroenterol. Hepatol. 2020;5:534–535. doi: 10.1016/S2468-1253(20)30118-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Liu Z., Chen Z., Huang X., Xu M., He T., Zhang Z. The establishment of reference sequence for SARS-CoV-2 and variation analysis. J. Med. Virol. 2020;92:667–674. doi: 10.1002/jmv.25762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Zhang Y., Zhang Z., Zhu J., Yu J. KaKs_Calculator 2.0: a toolkit incorporating Gamma-series methods and sliding window strategies. Dev. Reprod. Biol. 2010;8:77–80. doi: 10.1016/S1672-0229(10)60008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wang D., Zhang L., Sun W., Zhang Z., Chen W., Zhu A., Huang Y., Xiao F., Yao J., Gan M., Li F., Luo L., Huang X., Zhang Y., Wong S., Cheng X., Ji J., Ou Z., Xiao M., Li M., Li J., Ren P., Deng Z., Zhong H., Yang H., Wang J., Xu X., Song T., Mok C.K.P., Peiris M., Zhong N., Zhao J., Li Y., Li J., Zhao J. Intra-host variation and evolutionary dynamics of SARS-CoV-2 population in COVID-19 patients. bioRxiv. 2020 doi: 10.1186/s13073-021-00847-5. 2020.05.20.103549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood D.E., Lu J., Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20:257. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F., Tang M., Zheng X., Liu Y., Li X., Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158:1831–1833. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao M., Liu X., Ji J., Li M., Li J., Yang L., Sun W., Ren P., Yang G., Zhao J., Liang T., Ren H., Chen T., Zhong H., Song W., Wang Y., Deng Z., Zhao Y., Ou Z., Wang D., Cai J., Cheng X., Feng T., Wu H., Gong Y., Yang H., Wang J., Xu X., Zhu S., Chen F., Zhang Y., Chen W., Li Y., Li J. Multiple approaches for massively parallel sequencing of HCoV-19 (SARS-CoV-2) genomes directly from clinical samples. Genome Med. 2020;12:57. doi: 10.1186/s13073-020-00751-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Li X., Zhu B., Liang H., Fang C., Gong Y., Guo Q., Sun X., Zhao D., Shen J., Zhang H., Liu H., Xia H., Tang J., Zhang K., Gong S. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat. Med. 2020;26:502–505. doi: 10.1038/s41591-020-0817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Wen Y., Wang J., Gong Y., Feng K., Ye R., Jiang Y., Zhao Q., Pan P., Wu H., Duan S., Su B., Qiu M. The transmission and evolution of HIV-1 quasispecies within one couple: a follow-up study based on Next-generation sequencing. Sci. Rep. 2018;8:1404. doi: 10.1038/s41598-018-19783-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W.M., Song S.H., Chen M.L., Zou D., Ma L.N., Ma Y.K., Li R.J., Hao L.L., Li C.P., Tian D.M., Tang B.X., Wang Y.Q., Zhu J.W., Chen H.X., Zhang Z., Xue Y.B., Bao Y.M. The 2019 novel coronavirus resource. Yi Chuan. 2020;42:212–221. doi: 10.16288/j.yczz.20-030. [DOI] [PubMed] [Google Scholar]
- Zhou J., Li C., Liu X., Chiu M.C., Zhao X., Wang D., Wei Y., Lee A., Zhang A.J., Chu H., Cai J.P., Yip C.C-Y., Chan I.H-Y., Wong K.K-Y., Tsang O.T-Y., Chan K.H., Chan J.F-W., To K.K-W., Chen H., Yuen K.Y. Infection of bat and human intestinal organoids by SARS-CoV-2. Nat. Med. 2020:1–7. doi: 10.1038/s41591-020-0912-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Viral reads were deposited in the Genome Warehouse in the National Genomics Data Center (National Genomics Data Center Members and Partners, 2020) under project PRJCA2828, which are publicly accessible at https://bigd.big.ac.cn/gsa.