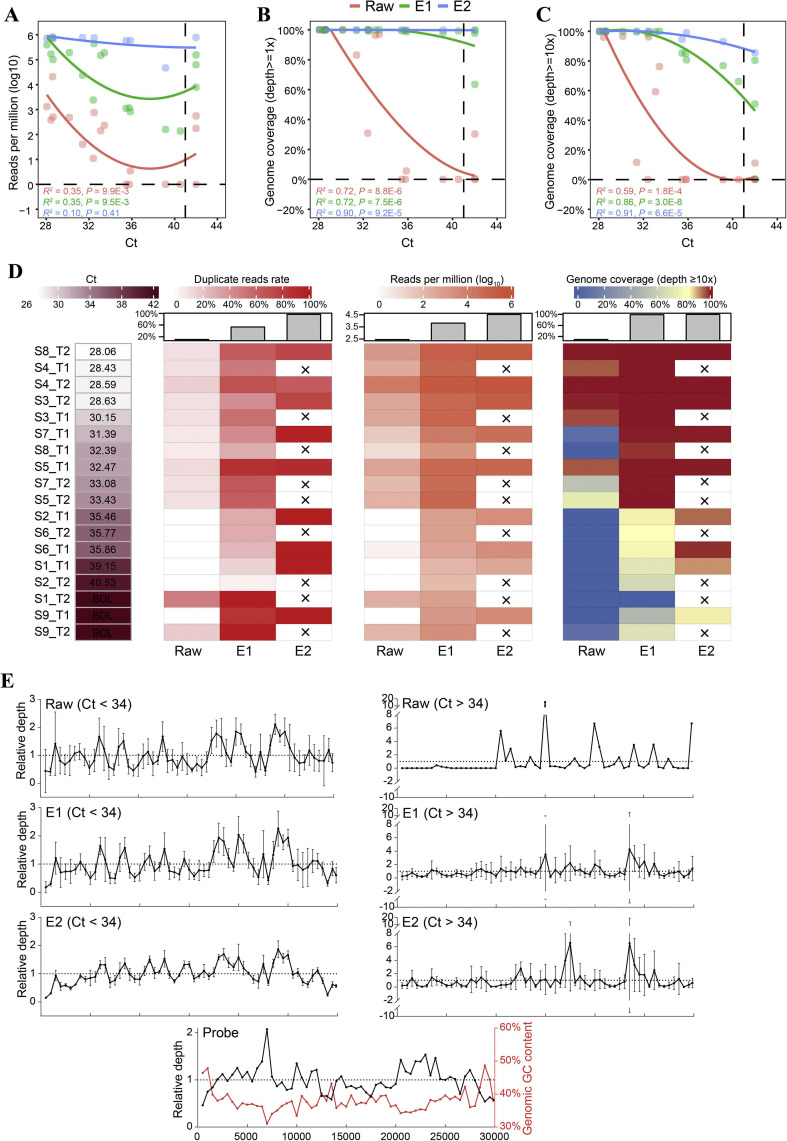
Fig. 1.
SARS-CoV-2 read counts and genome coverage obtained using direct metatranscriptomic sequencing and hybrid capture-based sequencing. A: The number of SARS-CoV-2 reads in the unit of reads per million (RPM). B: Genome coverage with depth ≥1. C: Genome coverage with and depth ≥ 10. In A–C, local polynomial regression line, R, and p values of Spearman correlations are shown; the Ct value that was below the detection limit was replaced with 42 for better visualization. D: Heatmap of Ct, duplicate reads rate, RPM, and genome coverage after deduplication. The median number for each group is shown with a bar plot, and crosses indicate that the samples were not included in E2 data. BDL, below the detection limit. E: Depth distribution of samples and probes along the SARS-CoV-2 genome. The relative depth was calculated by normalizing depth (bin size = 500 bases) to the average depth of each sample or the probes. Samples with Ct < 34 and Ct > 34 are shown separately; only samples with average depth > 1 are shown. The red curve indicates GC content along the reference genome. SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.