Summary
HIV-1 envelope (Env) N-glycosylation impact virus-cell entry and immune evasion. How each glycan interacts to shape the Env-protein-sugar complex and affects Env function is not well understood. Here, analysis of two Env variants from the same donor, with differing functional characteristics and N-glycosylation-site composition, revealed that changes to key N-glycosylation sites affected the Env structure at distant locations and had a ripple effect on Env-wide glycan processing, virus infectivity, antibody recognition, and virus neutralization. Specifically, the N262 glycan, although not in the CD4-binding site, modulated Env binding to the CD4 receptor, affected Env recognition by several glycan-dependent neutralizing antibodies, and altered site-specific glycosylation heterogeneity, with, for example, N448 displaying limited glycan processing. Molecular-dynamic simulations visualized differences in glycan density and how specific oligosaccharide positions can move to compensate for a glycan loss. This study demonstrates how changes in individual glycans can alter molecular dynamics, processing, and function of the Env-glycan shield.
Subject Areas: Biological Sciences, Biochemistry, Glycobiology, Microbiology, Virology
Graphical Abstract

Highlights
-
•
Two HIV-1 envelopes (Env) that differ in N-glycan composition were investigated
-
•
Changes in N-glycosylation had ripple effect on Env-wide glycan processing
-
•
Glycan changes impacted virus infectivity, antibody binding, and neutralization
-
•
These data revealed a functional role of glycan clusters in Env glycan shield
Biological Sciences; Biochemistry; Glycobiology; Microbiology; Virology
Introduction
The HIV-1 envelope glycoprotein (Env) is a trimer of gp160 proteins cleaved into two functional subunits: the gp41 trans-membrane glycoprotein and the exposed gp120 glycoprotein that recognizes CD4 receptor and CCR5 and/or CXCR4 co-receptors during virus-cell entry (Liu et al., 2008; Rizzuto et al., 1998; Wyatt et al., 1998). The multiple N-glycans of gp120 contribute to over 50% of the gp120 molecular mass (Kwong et al., 1998; Lee et al., 1992; Li et al., 1993; Pollakis et al., 2001), cover most of the Env protein surface, and are involved in key functions of HIV-1 biology (Bonsignori et al., 2012; Hessell et al., 2009; Krumm et al., 2016; Shivatare et al., 2018).
Analysis of single-genome-amplified sequences of viral RNA has identified nucleotide sequences of env genes from viruses responsible for productive HIV-1 infection (Abrahams et al., 2009; Keele et al., 2008), termed transmitted/founder viruses, as well as from viruses isolated during the chronic stage of infection of the same subjects (chronic-stage viruses) (Chun et al., 2013; Swanstrom and Coffin, 2012). HIV-1-infected subjects produce virus-neutralizing antibodies (nAbs), beginning approximately two months after appearance of virus-specific Abs (Davis et al., 2009; Tomaras et al., 2008). Generally, transmitted/founder viruses are sensitive to autologous nAbs (Liao et al., 2013; McCurley et al., 2017), which can impose selective pressure driving the emergence of mutated immune-escape variants. Sequence analysis of such variants suggests that the escape mechanism involves mutations in potential N-glycosylation sites (NGS) of Env gp120, resulting in loss of some potential NGS and appearance of new potential NGS (Richman et al., 2003; Wei et al., 2003). Consequently, the concept of an “evolving glycan shield” has been proposed (Richman et al., 2003; Wei et al., 2003) to explain the loss of neutralizing activity of the initial antibodies against newly emerging HIV-1 variants with altered composition and/or conformation of the Env glycans. Furthermore, the glycan shield has been recognized for its ability to protect from Env-specific nAbs even when N-glycans are not evolving (Kwong et al., 1998; Li et al., 1993; Pollakis et al., 2001; Stewart-Jones et al., 2016). However, the impact of these potential NGS mutations was only studied regarding their immediate location, and escape from nAbs, but not with respect to global effects on the Env structure. Our recent analysis of all deposited HIV-1 sequences in the LANL database demonstrated that certain N-glycan microdomains have a limited number of potential NGS combinations (Hargett et al., 2019). These findings suggest that some glycans in close proximity serve as clustered microdomains that have a distinct functional role in terms of position and glycan density to maintain the overall massive glycan shield and play a role in the proper folding of the Env structure (Hargett et al., 2019; Kong et al., 2015; Ozorowski et al., 2017).
In this study, we took advantage of two naturally occurring HIV-1 Env variants from the same donor that differed in functional characteristics and potential NGS composition and utilized them as a model system to define how glycans can affect the structure and function of Env. We have shown that changes to key potential NGS had a ripple-effect on Env-glycan processing at distant locations, with several glycans exhibiting altered processing. These site-specific alterations of Env glycans had impact on antibody recognition and interaction with CD4 receptor. Molecular-dynamic simulations enabled visualization of the complex changes induced by specific oligosaccharide positions and indicated that some glycans are able to move to compensate for a loss of a glycan and that glycan density (number of glycans within a region/microdomain) plays a functional role in the Env structure. These results confirm the interdependent nature of an NGS subset in the high-mannose patch that have been proposed to work as a microdomain of glycans (Hargett et al., 2019; Lemmin et al., 2017; Seabright et al., 2020). Together, these complementary applications of molecular biology, modeling, and functional glycomics revealed how individual glycans of the massive Env glycan shield can alter the Env glycan processing and, consequently, Env function.
Results
Selection and Characterization of HIV-1 Env with Distinct Glycosylation-Site Patterns
An essential prerequisite for studying glycan composition effects should be the identification of naturally occurring viral variants from a single donor with differing potential Env glycosylation-site composition and different functional characteristics. By using multiple-sequence alignment and NetNglyc software, we assessed WEAU HIV-1 Env from early and late stages of infection for encoded potential NGS (Fischer et al., 2010; Wei et al., 2003). We selected two Env variants with the most extreme differences in potential NGS, one from an early virus from day 16 after onset of symptoms of the acute retroviral syndrome (designated WEAU-d16) and the other from a late virus from day 391 (designated WEAU-d391) for further analyses. Env gp120 of WEAU-d16 and WEAU-d391 exhibited seven differentially positioned potential NGS in the V1, C2, the C2-V3 junction, and V4 regions (Figures 1A, 1B, and S1); three unique potential NGS (N141, N295, and N398) were detected in WEAU-d16 and four unique potential NGS (N234, N262, N396, and N406) in WEAU-d391.
Figure 1.
Differences in Potential N-glycosylation Sites (NGS) of WEAU gp120 and Infectivity of WEAU Early Virus (WEAU-d16) and Chronic-Stage Virus (WEAU-d391)
(A) Potential NGS determined by amino acid motifs N-X-S/T (N, Asn; S, Ser; T, Thr) in gp120 of WEAU-d16 and WEAU-d391. Abbreviation: PNGS, potential N-glycosylation sites; C, constant regions; V, variable regions.
See also Figure S1.
(B) In silico comparative analysis of WEAU-d16 and WEAU-d391 gp120 potential NGS performed by http://www.cbs.dtu.dk/services/NetNGlyc/. The threshold of likely glycosylation was set at 0.25 and the predicted glycosylation sites are marked by a vertical line. Seven unique potential NGS are marked by blue (WEAU-d16) and red (WEAU-d391) vertical lines. Constant (C) and variable (V) regions are marked.
(C) Infectivity of WEAU Env-pseudotyped viruses produced in 293F cells using TZM-bl cells (left) or J2574 reporter T cells (right). Virus infectivity using TZM-bl reporter cells is expressed in relative light units (RLU) normalized to virus load (quantified by p24 content). Virus infectivity using J2574 reporter T cells is expressed as percentage of GFP-positive cells normalized to virus load (p24).
See also Figures S2 and S9.
Data information: In panel C, data from eight TZM-bl and three J2574 independent experiments are shown as means ± standard deviations. p values calculated by using two-tailed Student's t test: ∗∗∗p < 0.0000001; ∗∗p < 0.001.
Functional Characterization of the Selected HIV-1 WEAU Variants
To assess the functional impact of differential potential NGS in the selected WEAU Envs, we generated synthetic gp160 genes of WEAU-d16 and WEAU-d391 and produced WEAU Env-pseudotyped viruses. These gp160 genes had identical gp41 segments (derived from WEAU-d16), whereas gp120 genes, based on WEAU-d16 sequence, reflected the strain-specific potential NGS (Figure S1). The infectivity of WEAU-d16 virus was significantly lower compared with WEAU-d391 using TZM-bl reporter cells (Montefiori, 2009) (Figure 1C, left; Figure S2) and J2574 reporter T cells (Jones et al., 2007) (Figure 1C, right).
Differential infectivity was not the result of potential artifacts in the production of Env-pseudotyped viruses, as revealed by western blotting with serum IgG from an HIV-1-infected subject. The viral protein content and processing, including uncleaved Env (gp160), cleaved Env (gp120, gp41), p24, and other proteins, were similar for WEAU-d16 and WEAU-d391 virions (Figures 2A, 2B, and S3).
Figure 2.
Virions with Env from an Early Virus (WEAU-d16) and Chronic-Stage Virus (WEAU-d391) Exhibit Similar Protein Components, Including Env Expression, Incorporation, and Cleavage
(A) Representative western blot assays of 20 times concentrated viral stocks (two preparations each). Nine HIV-1 proteins were detected using serum IgG antibodies from an HIV-1-infected individual. The amounts of uncleaved Env (gp160), cleaved Env (gp120), and p24 proteins were quantified by densitometry as integrated intensity (i.i.) of each protein band.
(B) Env incorporation into virions was calculated using the formula (gp160 i.i. + gp120 i.i.)/p24 i.i. and the Env cleavage (gp120) was calculated using the formula gp120 i.i./(gp120 i.i. + gp160 i.i.). Virus infectivity was determined in eight independent experiments using TZM-bl reporter cells and expressed in relative light units (RLU) normalized to virus load (p24).
See also Figure S3.
The N262 Glycans Affect Binding to CD4 and Recognition by CD4-Binding-Site-Targeting Broadly Neutralizing Antibodies in Native as well as Recombinant gp120 Trimers
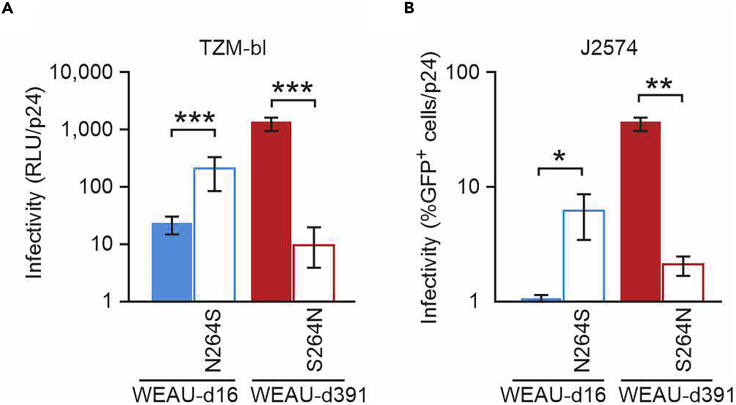
Among the seven distinct potential NGS between WEAU-d16 and WEAU-d391, we were particularly interested in the N262 NGS, the site present in WEAU-d391 but not in WEAU-d16 (Figures 1A and 1B). This site, previously shown to affect HIV-1 infectivity (Mathys et al., 2014; Wang et al., 2013), is not located in any Env domain with a designated direct functionality (e.g., CD4-binding site), but rather involved in gp120 folding and stability (Kong et al., 2015). To test whether the low infectivity of WEAU-d16 may be due to lack of the N262 glycans, we generated WEAU-d16 Env with the N262 NGS added (N264S; designated WEAU-d16-N264S) and WEAU-d391 with N262 NGS removed (S264N; designated WEAU-d391-S264N). Indeed, WEAU-d16-N264S Env-pseudotyped virus exhibited 9- and 7-fold higher infectivity in TZM-bl and J2574 reporter cells compared with the parental WEAU-d16 virus (Figures 3A and 3B). Conversely, WEAU-d391-S264N virus exhibited a significantly reduced infectivity compared with the parental WEAU-d391 virus—100- or 20-fold reduced infectivity of TZM-bl or J2574 reporter cells, respectively (Figures 3A and 3B).
Figure 3.
Infectivity of Virions Pseudotyped with Env from an Early Virus (WEAU-d16) and Env from Chronic-Stage Virus (WEAU-d391) is Impacted by the Presence of N262 Glycan
(A) Viral infectivity determined using TZM-bl reporter cells is expressed in relative light units (RLU) normalized to virus load (p24).
See also Figure S9.
(B) Infectivity using J2574 reporter T cells is expressed as percentage of GFP-positive cells normalized to virus load (p24). WEAU-d16-N264S is a WEAU-d16 mutant with N262 potential NGS introduced. WEAU-d391-S264N is a WEAU-d391 mutant without N262 potential NGS. The WEAU viruses without N262 potential NGS had significantly lower infectivity.
Data information: Data from eight TZM-bl and three J2574 independent experiments are shown as means ± standard deviations. p values are calculated by using two-tailed Student's t test: ∗∗∗p < 0.001; ∗∗p < 0.01; ∗p < 0.05.
To define whether the loss of infectivity associated with the absence of the N262 NGS was related to altered virus-CD4 binding, we tested binding of WEAU Env-pseudotyped viruses to recombinant soluble CD4 (sCD4). Infectivity of viruses containing the N262 NGS was inhibited by sCD4 in a dose-dependent manner (Figures 4A and 4B). The IC50 was 2.46 ± 0.21 μg/mL for WEAU-d391 and 1.43 ± 0.42 μg/mL for WEAU-d16-N264S. Inhibition by sCD4 was approximately 4-fold lower (measured as IC25) when N262 NGS was removed from WEAU-d391 (Figures 4A and 4B). Similarly, inhibition by sCD4 was approximately 6-fold lower for WEAU-d16 (Env without N262 NGS) compared with the mutant with N262NGS added (WEAU-d16-N264S). These results suggest that, although N262 NGS is not located in the CD4-binding site, the N262-associated glycan controls the Env conformation in regard to CD4-binding capacity.
Figure 4.
Soluble CD4 Inhibits Infectivity of WEAU Env-pseudotyped Viruses with N262 NGS More Than Those Without N262 NGS
(A) Mean and SD values for serial dilutions of sCD4 from three independent experiments.
(B) Mean and SD values were calculated from three independent experiments for IC50 and IC25.
Data information: Data from three independent experiments are shown as means ± standard deviations for each dilution (A) and IC50 and IC25 (B). 262NGS indicates N262 NGS presence (WEAU-d391 and WEAU-d16-N264S), and Δ262NGS indicates N262 NGS absence (WEAU-d16 and WEAU-d391-S264N).
To further clarify the function of the N262 glycans, we produced recombinant trimeric gp120 WEAU variants using our previously published approach that added a foldon and V5 and His tags to the gp120 (Hargett et al., 2019; Raska et al., 2008, 2010, 2014). We confirmed that gp120 WEAU glycoproteins were exclusively produced as trimers (Figure 5A).
Figure 5.
Binding of Soluble CD4 and CD4-Binding-Site-Specific Broadly Neutralizing Antibodies (BnAbs) to Recombinant gp120 Trimers is Enhanced by the Presence of N262 Glycan
(A) Recombinant proteins WEAU-d16, WEAU-d391, and a reference gp140 trimer (HIV-1SF162) (at 2, 1, and 0.5 μg protein) were fractionated using a 4% to 15% polyacrylamide gradient Mini-PROTEAN TGX precast gel (Bio-Rad) under native conditions. Protein bands ~420 kDa visualized using Bio-Safe Coomassie blue G-250 stain indicated that the preparations are forming trimers under native conditions.
(B) The binding of recombinant gp120 trimers (rgp120) to soluble CD4 (sCD4) was measured using sCD4-coated ELISA plates and expressed as optical density at 490 nm (OD490) normalized to rgp120 protein.
(C and D) Equal amounts of rgp120 were added to ELISA plates coated with mouse anti-His antibody. Serially diluted samples of VRC01 (C) or b12 (D) BnAb were then added, and Ab binding was expressed as OD490 values. 262NGS indicates N262 potential NGS presence (WEAU-d391 and WEAU-d16-N264S) and Δ262NGS indicates N262 potential NGS absence (WEAU-d16 and WEAU-d391-S264N).
Data information: For panels B, C, and D, data from two experiments are shown as means ± standard deviations.
The four trimeric recombinant WEAU gp120 (rgp120) variants were analyzed by ELISA and western blotting to determine rgp120 protein concentrations (Raska et al., 2014). rgp120 WEAU-d16 and WEAU-d391-S264N exhibited low level of binding to ELISA-coated sCD4, whereas binding of rgp120 WEAU-d16-N264S and WEAU-d391 was 2- to 4-fold higher (Figure 5B). These results reproduced the data obtained for sCD4-mediated inhibition of virus infectivity with the respective WEAU Env variants (Figure 4).
The trimeric rgp120 variants also allowed us to test whether the N262 NGS affected recognition of the CD4-binding site by the well-characterized broadly neutralizing antibodies (BnAbs) b12 and VRC01. Both BnAbs exhibited strong binding to rgp120 variants with the N262 NGS (WEAU-d16-N264S and WEAU-d391) at the range of concentrations tested (0.0625–2 μg/mL) (Figures 5C and 5D). Conversely, b12 did not bind to and VRC01 had a reduced ability to recognize rgp120 variants without N262 NGS (WEAU-d16 and WEAU-d391-S264N).
Together, these data demonstrate that the N262 glycans, although not located in the CD4-binding site, are crucial for Env-CD4 binding and viral infectivity. Essential for the ensuing glycomics studies, the data demonstrate that the corresponding structural and functional characteristics of native Env, as expressed on the virus, are preserved in the recombinant Env variants.
The N262 N-Glycosylation Site Impacts the Protein-wide Glycan Composition of rgp120 Trimer in a Site-Specific Context
Given the effect of the N262 glycans on Env structure and functionality, we decided to decipher how the N262 glycans could globally affect the Env glycan shield. To begin to address this question, we performed monosaccharide compositional analysis by using gas-liquid chromatography to assess the global glycan composition of WEAU rgp120 trimer variants (Hargett et al., 2019; Raska et al., 2010, 2014). Surprisingly, the monosaccharide compositional analysis revealed that the presence of N262 not only affected function but also impacted the overall glycan composition of the Env gp120 trimers. Env variants with the N262 potential NGS (WEAU-d391 and WEAU-d16-N264S) had a lower galactose content compared with Env variants without N262 potential NGS (WEAU-d16 and WEAU-d391-S264N). Galactose to mannose ratios were 0.51 and 0.42 for WEAU-d16 and WEAU-d16-N264S and 0.54 and 0.64 for WEAU-d391 and WEAU-d391-S264N. As galactose is present in most complex glycans, but absent from high-mannose glycans, this observation suggested overall enhanced glycan processing toward complex glycans in trimeric rgp120 variants without N262 potential NGS.
To determine the specific nature of changes at the individual potential NGS, we used our validated LC-MS-based glycomics workflow (Hargett et al., 2019). Trimeric rgp120 preparations with N262 NGS (WEAU-d16-N264S and WEAU-d391) and without N262 potential NGS (WEAU-d16 and WEAU-d391-S264N) were normalized for total protein (Figure S4), digested with various combinations of proteases, and the subsequent (glyco)peptides were analyzed by LC-MS. These results are in agreement with our previous analysis and provided an opportunity to further examine changes in site-specific glycan heterogeneity (Hargett et al., 2019). The glycopeptides for each NGS were expressed as relative abundance of each individual N-glycopeptide at each site for each rgp120 variant. Three different types of heterogeneity were observed that categorized individual NGS as (1) predominantly high-mannose glycans (>55%), (2) predominantly complex glycans (>55%), and (3) a mixed population of both high-mannose and complex glycans (i.e., high mannose and complex glycans each comprised <50% of total glycan distribution) (Figures 6A and S4) (Hargett et al., 2019). N-glycan site-specific heterogeneity profiles of the four WEAU rgp120 trimers are shown in Figure 6A compared with those published for other gp120 trimers (Behrens et al., 2017). As expected, we found N262 glycosylated in WEAU-d391 and WEAU-d16-N264S rgp120 variants, with the site occupied by a mixture of high-mannose and complex glycans.
Figure 6.
Cross-Sample Comparison of Site-Specific Glycosylation Heterogeneity of Four Different WEAU gp120 Trimers Reveals the Impact of N262 Glycan on Heterogeneity of Several Other Spatially Adjacent Glycans
(A) Site-by-site comparison of the glycosylation profiles of four WEAU gp120 trimers (WEAU-d16, WEAU-d391, WEAU-d16-N264S, WEAU-d391-S264N) compared with those of BG505 gp120 monomer and two BG505 trimers (afrom reference (Go et al., 2017)). To enable presentation of sites across all gp120 variants, we followed the style and nomenclature reported by (Go et al., 2017). Sites are classified as mostly high-mannose (H), mostly processed (C), or mixture of high-mannose and processed (M). Some sites were not detected in the analysis (ND), and some sites were not present in a given variant (X). Some NGS are observed as a single glycopeptide with two glycosylated sites in the LC-MS analyses (i.e., N188-N197) in ours and the reported analyses by Go et al. (Go et al., 2017). Some sites are labeled to reflect different sites of glycan attachment in different Env variants (e.g., N355-N356, N411-N413); for details on NGS in WEAU variants, please see Figures 1 and S1. N262 and the glycosylation sites shown in panels B–D are highlighted with pink-colored background.
(B) Comparison of abundance of all high-mannose glycans at three representative NGS: N448 (variable N-glycan composition in different WEAU gp120 trimers), N413a (predominantly high-mannose N-glycans), and N276 (predominantly processed N-glycans). There was significantly more high-mannose glycans at N448 in gp120 trimers with N262 NGS than in those without N262 NGS.
(C) Some NGS demonstrate significant changes within the high mannose category of N-glycans between variants. The ratio of the Man9 oligosaccharide (HM9) to the Man5 oligosaccharide (HM5) from quantitative site-specific glycan profiling for three predominantly high-mannose-containing NGS. N413a contained significantly more Man9 glycans than Man5 in gp120 trimers with N262 NGS than in those without N262 NGS. N332 showed minor differences, and N339 had significantly more Man9 glycans in the WEAU-d391 gp120 trimer compared with the other three gp120 trimer variants.
(D) The ratio of Hex3HexNAc4Fuc1 oligosaccharide to Hex5HexNAc4Fuc1 oligosaccharide for predominantly processed N-glycans. N88 NGS and N160 NGS had significantly more Hex3HexNAc4Fuc1 than Hex5HexNAc4Fuc1 for the WEAU-d391 gp120 trimer, implying less processing of individual glycans in some NGS for this gp120 trimer variant. Color coding for panels in B–D is shown at the bottom, below panel (D).
(E) The glycan heterogeneity profile of N276 contains predominantly complex glycans (blue). WEAU-d391 gp120 trimer has ~15% less complex glycan compared with the other gp120 trimer variants.
See also Figure S5.
Data information: For panels B, C, and D, data are represented as mean ± SEM.
To determine the effect of N262 NGS on other Env glycans, quantitative differences in the site-specific heterogeneity of individual NGS were assessed at the overall classification level (relative proportion of high-mannose, hybrid, and complex glycans) as well as at the representation of individual oligosaccharides. Of the sites that were common among all four WEAU Env variants, N448 exhibited the greatest differences in its overall N-glycan profile. In Env variants with N262 NGS (WEAU-d16-N264S and WEAU-d391), N448 NGS had >65% high-mannose glycans (Figure 6B). In Env variants without N262 NGS, representation of high-mannose glycans at N448 was reduced by 20% (WEAU-d16-N264S versus WEAU-d16) and by 30% (WEAU-d391 versus WEAU-d391-S264N). A similar trend was seen for the glycans at N241 (Figure 6A). The remaining NGS exhibited relatively small differences (<10% shift) in high-mannose-glycan content across the WEAU variants (Figure 6A). These NGS followed the quantitative trends shown in Figure 6B for the predominantly high-mannose at N413a NGS and the predominantly processed glycans N276 NGS.
Most NGS of the four WEAU rgp120 variants did not differ in their category of single-site heterogeneity (i.e., predominantly high-mannose or complex glycans or mixture of both types). Instead, several NGS showed differences within the same category of glycans (e.g., different proportion of Man9GlcNAc2 to Man5GlcNAc2 high-mannose glycans). The first steps in N-glycan processing include the conversion of Man9GlcNAc2 into Man5GlcNAc2. In Figure 6C, we compared the ratio of Man9GlcNAc2 oligosaccharide with that of the Man5GlcNAc2 oligosaccharide at NGS N413a, N332, and N339. N413a NGS, a site that is proximal to N262 (Stewart-Jones et al., 2016), had more Man9GlcNAc2 than Man5GlcNAc2 in rgp120 variants with N262 NGS, indicating less processing at N413a NGS in WEAU rgp120 trimers with N262 NGS versus those without N262 NGS. In contrast, similar ratio of these two high-mannose glycans (Man9GlcNAc2 and Man5GlcNAc2) at N332 in all four WEAU variants (also proximal to N262) showed little impact of N262 NGS. Interestingly, NGS N339 in the WEAU-d391 rgp120 trimer had significantly more Man9GlcNAc2 than Man5GlcNAc2 when compared with the other three WEAU rgp120 variants (Figure 6C).
The remaining NGS in our glycomics analysis showed little global change in single-site heterogeneity. However, there was a consistent pattern within the complex glycans that indicated a difference in the extension of complex glycans in the WEAU-d391 rgp120 trimer versus other trimers. The quantitative ratio of two complex glycans, Hex3HexNAc4Fuc1 and Hex5HexNAc4Fuc1, at NGS with complex glycans (N88, N160; Figure 6D) showed a consistently higher abundance of Hex3HexNAc4Fuc1. This observation suggested that N-glycan shield of the WEAU-d391 variant was the least processed of the WEAU trimers analyzed, even at NGS that primarily contain complex N-glycans. This conclusion was further reflected in the overall N-glycan heterogeneity of N276 where the WEAU-d391 rgp120 trimer was the only variant that had ~50% complex glycans, a difference of ~15% compared with the other three trimers (Figure 6E).
Thus, these high-resolution quantitative glycomics results (Figures 6 and S5) (Hargett et al., 2019), examined in the context of the functional analysis, detail how a local, single glycosylation site (N262 NGS) influenced the N-glycan heterogeneity of rgp120 trimers and exert a glycomic ripple effect that potentially propagates functional and conformational changes at local and distal sites throughout the Env trimer.
Modeling the Impact of Positional Glycosylation Differences on Global Env Structure
The glycomic profiling detailed above provided empirical evidence of the changes imparted by N262 NGS. To begin to understand the implications of these changes, we mapped the glycoprofiling data of WEAU variants onto the structure of the gp120 trimer. The observed differences confirm a cluster of interdependent NGS spatially close to N262. We previously proposed this working cluster unit (Hargett et al., 2019) as a glycan microdomain within the so-called “high-mannose patch” (Pritchard et al., 2015) that in these variants consists of N262, N295, N301, N332, N413a, and N448 NGS (high-mannose patch [HMP]-microdomain). This same glycan cluster has been proposed by structural analysis based on proximity and modeling simulations that show interaction of these NGS within the high-mannose patch (Lemmin et al., 2017). The NGS variations in this region among the four studied Env variants empirically confirm these interactions. Together, these results provide an insight into the role of this glycan cluster in the overall structure of the Env trimer.
To visualize how differences in this glycan cluster influence the entire Env trimer, we performed a series of molecular dynamics simulations (MDS) of the two WEAU variants and the corresponding N262 mutants. By using the solved crystal structure of HIV-1 clade B JR-FL prefusion Env trimer as a template (PDB: 5FYK) (Stewart-Jones et al., 2016), homology models were generated for all four WEAU variants (WEAU-d16, WEAU-d16-N264S, WEAU-d391, and WEAU-d391-S264N). Man5GlcNAc2 oligosaccharides were modeled at each sequon using an in-house glycosylator software. To analyze glycan motions in different WEAU trimers, 500-nanosecond (ns) MDS were performed (Videos S1, S2, S3, and S4). Figure 7, showing initial glycan positioning for MDS within the HMP-microdomain of Env gp120, illustrates different glycan densities of the clustered glycans of this region in the four WEAU Env variants. A qualitative assessment of WEAU-d391-S264N MDS, the variant with the lowest glycan density within the high-mannose patch, revealed N301, N332, N413a, and N448 oligosaccharides to exhibit relatively free movements. Notably, the protein cleft normally occupied by glycan N262 was vacant. As the number of glycans within the HMP-microdomain increased by one glycan for WEAU-d16 and WEAU-d391 or by two glycans for WEAU-d16-N264S, movement of oligosaccharides became more restricted in these trimers. These findings indicate that glycan density in this HMP-microdomain differentially impacts glycan movements and that specific glycans, especially N262, can have large effects with functional implications.
Figure 7.
Initial Glycan Positioning for MDS with the gp120 High-Mannose Patch on Envelope Trimers WEAU-d16, WEAU-d16 N264S, WEAU-d391, and WEAU-d391-S264N
(A–D) Glycan conformations are modeled onto the HIV-1 envelope trimer molecule using an in-house glycosylator software that arranges glycans avoiding atom clashes. Selected glycans are highlighted in stick conformation for (A) WEAU-d16, (B) WEAU-d16-N264S, (C) WEAU-d391, (D) WEAU-d391-S264N.
Glycans are marked by numbers and color coded as in the Supplemental Videos: Orange: N262 (if present), Blue: N295 (if present), Red: N301, Yellow: N332, Green: N413, Pink: N448.
Based on the initial qualitative assessment of the MDS for the four WEAU variants, we next performed a network analysis on the dense array of glycan-glycan interactions. We used changes in betweenness centrality (Kwong et al., 1998; Li et al., 1993; Pollakis et al., 2001; Stewart-Jones et al., 2016), a measure representing the number of shortest paths that pass through a node, as a means to identify changes in the interactions between the glycans. A heatmap of the betweenness centrality for each oligosaccharide was generated and overlaid onto each WEAU model (Figure S6). When all WEAU variants were compared, we observed no differences in the betweenness centrality at the CD4-binding site. However, the extent of betweenness centrality varied among protomers in the HMP-microdomain and the trimer apex. At protomer 2, there is a clear path of betweenness centrality from the base of the trimer to the apex in all WEAU variants. At protomer 3, the WEAU-d391-S264N high-mannose patch contained little or no betweenness centrality, whereas all other variants had a high degree of betweenness centrality. At protomer 1, the high-mannose patch of WEAU-d16 and WEAU-d391-S264N contained disconnected regions of betweenness centrality, whereas WEAU-d391 and WEAU-d16-N264S exhibited a continuous trail of glycans with a higher betweenness centrality from the apex of the trimer to the base. This included a degree of betweenness centrality between N448 and N241, which likely explains the shift in N241 heterogeneity seen in our glycomics analysis when N262 was absent. Thus, these features differentiated the variants with N262 glycan (WEAU-d391, WEAU-d16-N264S) from those without the N262 glycan (WEAU-d16, WEAU-d391-S264N). These data strongly support the functional and glycomics data regarding the ripple effect of N262 glycan on the entire Env trimer.
Next, we averaged the location of the center of mass (COM) for each oligosaccharide spatially close to the N262 NGS over the 500-ns MDS. To determine the effect the N262 glycan had on the position of the neighboring glycans, we determined the differences in the COM locations of those glycans in WEAU-d16-S264N (with N262) and WEAU-d16 (without N262) for all three protomers of the HIV-1 Env trimers (Figures 8 and S7). For protomer 2, N413a shifted 12.52 Å toward N295 in WEAU-d16-S264N compared with WEAU-d16. N295 shifted 11.75 Å toward N448 in WEAU-d16-S264N. N448 shifted 7.71 Å toward N262 in WEAU-d16-S264N. N301 and N332 position changed only slightly (2.92 Å and 3.57 Å, respectively) (Figure 8A). We then determined the differences in the COM locations in WEAU-d391 (with N262) and WEAU-d391-S264N (without N262) (Figure 8B). The N448 glycan shifted 5.63 Å toward N262. N332 shifted 6.23 Å away from the β-sheet. N301 and N413a shifted only 4.24 Å and 0.81 Å, respectively. Interestingly, the COM of N413, N295, and N448 glycans in WEAU-d16 shifted to compensate for the loss of the N262 oligosaccharide. Conversely, in WEAU-d391, only the COM of N448 shifted toward the COM of N262 (Figure S7). The COM shifts for protomer 1 and protomer 3 for the HIV-1 Env trimers showed a similar trend (Figure S7). This observation indicates that the lack of glycan density in the WEAU-d391-S264N created less steric push to fill the N262 void.
Figure 8.
Center of Mass (COM) Data for Glycans within the High-Mannose Patch Highlight Glycan Movement When Glycan N262 is Absent
(A) COM differences were calculated for each oligosaccharide between WEAU-d16-N264S (dark green) and WEAU-d16 (purple) for protomer 2. The resultant COM shifts were observed: 2.9 Å for N301, 3.6 Å for N332 Å, 11.8 Å for N295, 12.5 Å for N413, and 7.7 Å for N448.
(B) COM differences were calculated for each oligosaccharide between WEAU-d391 (purple) and WEAU-d391-S264N (dark green) for protomer 2. The resultant COM shifts were observed: 4.3 Å for N301, 6.2 Å for N332 Å, 0.8 Å for N413, and 5.6 Å for N448. N262 NGS in WEAU-d16-N264S and WEAU-d391 is shown in orange.
See also Figures S6 and S7.
Overall, the MDS enabled visualization of the impact of the N262 glycan on HIV-1 Env N-glycan shield. The resultant weaker glycan-glycan interactions were manifested by the altered betweenness centrality plots for WEAU-d391-S264N protomer 1 and 3 and WEAU-d16 protomer 3. Furthermore, the COM calculations revealed how specific oligosaccharide positions can move to compensate for the loss of the N262 glycan, thereby altering the N-glycan shield in this region of the Env trimer.
N262 NGS Impacts Neutralization of HIV-1 by Glycan-dependent/co-dependent BnAbs
Our functional glycomics and MDS data suggested that the variability in the cluster of glycans surrounding N262 impacted several sub-regions of the glycan shield of the Env trimer. In addition, in our previous analysis of these variants, we evaluated the frequency of the NGS sequon combinations in this N262-impacted HMP-microdomain. The two HMP-microdomain NGS variants used in our studies are representative of nearly 50% of all sequences in the LANL database in terms of this HMP-microdomain. Several of the HMP-microdomain NGS are critical for the interaction of some BnAbs with Env. Thus, if our hypothesis of the ripple effect of N262 were true, regional changes in the glycan shield could be observed through a panel of BnAbs that have known glycan interactions. If correct, the functional implications of the observed dynamic changes in gp120 glycan shield should result in an altered ability of BnAbs to bind to the N262 Env variants. We thus tested neutralization sensitivity of HIV-1 virions pseudotyped with Env variants with N262 NGS (WEAU-d391 and WEAU-d16-N264S) and without N262 NGS (WEAU-d16 and WEAU-d391-S264N) to BnAbs with confirmed glycan-(co)dependent activity (Table 1 and Figure S8).
Table 1.
Neutralizing Activity of Some BnAbs against WEAU Pseudotyped Viruses with Env Variants with and without PNGS N262 Is Impacted by N262 Glycan
| BnAbs | Regions/Epitope Features | IC50 (μg/mL) |
Fold Changea,b |
||||
|---|---|---|---|---|---|---|---|
| WEAU-d16 |
WEAU-d391 |
WEAU-d16 |
WEAU-d391 |
||||
| N262 | ΔN262 | N262 | ΔN262 | N262/ΔN262 | N262/ΔN262 | ||
| 2F5 | gp41/N-MPERf ELDKWAS | 0.562 | 0.335 | 0.725 | 0.35 | 1.7 | 2.1 |
| 4E10 | gp41/C-MPERg NWFDITN | 1.4 | 0.401 | 1.3 | 1.2 | 3.5 | 1.1 |
| PGT151 | gp120-gp41 IFe/glycans N611, N637 | 1.592 | 0.093 | 0.714 | 0.013 | 17.1 | 54.9 |
| 35O22 | gp120-gp41 IF/glycan N88 | 0.18 | 0.024 | 0.323 | 0.048 | 7.5 | 6.7 |
| 2G12 | C2 C3 V4 CD4bsd/glycan patch N295, N332, N386, N392, N448 | 1.4 | 0.194 | >33.3 | >33.3 | 7.2 | >33.3/>33.3 |
| VRC24 | V3/glycan N301 | 3.0 | 0.033 | 12.7 | 0.131 | 90.9 | 96.9 |
| PGT128 | V3 C3/glycans N301, N332 | 0.039 | 0.008 | 0.056 | 0.015 | 4.9 | 3.7 |
| PGT121 | C3/glycan N332 | 0.029 | 0.019 | 0.014 | 0.011 | 1.5 | 1.3 |
| PGT122 | C3/glycan N332 | 0.055 | 0.045 | 0.085 | 0.041 | 1.2 | 2.1 |
| PGT135 | C3 V4/glycans N332, N386, N392 | 0.361 | 0.149 | 0.341 | 0.108 | 2.4 | 3.2 |
| PG9 | V1 V2/glycans N156 or N173, N160 | 3.7 | 4.3 | 10.5 | 0.243 | 0.9 | 43.2b |
| PG16 | V1 V2/glycans N156 or N173, N160 | >33.3c | 3.6 | 12.2 | 0.819 | >9.3 | 14.9 |
| PGT145 | V2/glycan N160 | 1.9 | 0.112 | 11.2 | 0.24 | 17.2 | 46.7 |
| VRC01 | CD4bs | 0.076 | 0.051 | 0.139 | 0.072 | 1.5 | 1.9 |
| b12 | CD4bs | 0.656 | 0.228 | 1.6 | 0.786 | 2.3 | 2.0 |
Ratio of IC50 values; see also Figure S8.
Fold change indicates IC50 of WEAU N262/IC50 WEAU ΔN262. Values with fold differences >5 are in bold font.
Less than 50% inhibition of viral infection was determined with the highest input of antibody (33.3 μg/mL) in the assay.
CD4bs: CD4-binding site.
IF: interface.
N-MPER: N-terminal membrane proximal external region.
C-MPER: C-terminal membrane proximal external region.
BnAbs specific for gp41 (2F5, 4E10) showed no or only small differences in their capacity to neutralize viruses with WEAU Env variants with and without N262 NGS, thus serving as a control of where the glycan shield was not altered by N262 removal. In contrast, PGT151 and 35O22, BnAbs that interact within the gp120-gp41 interface (Blattner et al., 2014; Falkowska et al., 2014; Huang et al., 2014; Lee et al., 2015), exhibited differences in virus-neutralization capacity, based on N262 presence or absence. Specifically, PGT151, which has an overlapping epitope with N448, N611, N616, and N637 NGS (Lee et al., 2016), exhibited >50-fold better neutralization (based on IC50 values) of WEAU-d391-N264S compared with WEAU-d391 viruses (Table 1). Although N448 was present in all four tested Env variants, its position and glycan composition were significantly altered by the loss of N262.
Several BnAbs with glycan-dependent binding within the V3, C3, and V4 regions of gp120 target glycan epitopes that are part of our proposed N262-anchored glycan cluster. These BnAbs exhibited differences in neutralization activity of the tested WEAU viruses. For example, 2G12, BnAb that targets conformational epitopes consisting of high-mannose glycans (Hessell et al., 2009; Murin et al., 2014), neutralized only WEAU-d16 and WEAU-d16-N264S but not WEAU-d391 and WEAU-d391-S264N. This resistance of the immune-escape WEAU-d391 virus to 2G12 neutralization was likely due to the absence of N295 potential NGS, one of the glycans forming the 2G12 epitope (Wei et al., 2003). WEAU-d16-N264S was 7-fold less sensitive to neutralization by 2G12 than WEAU-d16, based on IC50 values. Although the WEAU-d16-N264S mutant still had N295, it also had N262 to anchor this cluster of glycans. Notably, serum IgG from WEAU subject five years after acquiring the infection still contained nAbs against early virus variant (WEAU-d16) but not the chronic-stage immune-escape variant (WEAU-d391, gained N262, lost N295) that was also resistant to BnAb 2G12 (Figure S8). VRC24, BnAb interacting with V3 and N301 NGS (Georgiev et al., 2013), exhibited >90-fold better neutralization (based on IC50 values) of WEAU virions without N262 NGS than those with N262 NGS (Table 1).
Conversely, PGT128, BnAb targeting the epitope in the V3 loop and the N-glycans at N301 and N332 (Doores et al., 2015; Pejchal et al., 2011), exhibited similar neutralization capacity against all four WEAU variants. This observation is in agreement with the previous finding that for Env variants with N332 NGS, the N301 glycan becomes nonessential for neutralization by PGT128-type BnAbs (Krumm et al., 2016). As expected, other BnAbs that interact with N332 glycan (Krumm et al., 2016), such as PGT121, PGT122, and PGT135, also exhibited similar neutralization capacity against the tested WEAU viruses irrespective of N262 NGS. These results imply N301 is more dependent upon N262-related interactions than N332.
PG9, PG16, and PGT145, BnAbs that target the apex of the Env trimers, exhibited differences in neutralization of WEAU pseudotyped viruses dependent on N262 NGS. PG9 and PG16 recognize glycan-containing regions of V1/V2, including glycans at N156 or N173 and N160 (Euler et al., 2011; McLellan et al., 2011). PG9 exhibited >40-fold higher neutralization capacity for WEAU-d391 without versus with N262 NGS. However, similar neutralization capacity was observed for WEAU-d16 variants irrespective of N262 NGS. This suggests that PG9 neutralization is dependent upon N262 in some variants, but WEAU-d16 is susceptible to PG9 neutralization. In contrast, both PG16 and PGT145 exhibited higher neutralization capacity for WEAU variants without N262 NGS, implying their neutralization capacity is affected by the role of N262 in the formation of the apex of the Env trimers. Similarly, BnAbs specific for CD4-binding site, VRC01 and b12, exhibited a slightly better neutralization of virions without N262 glycans (Table 1). These results are in contrast with the results of inhibition of viral entry by sCD4, where the inhibition impacted the virions with N262 glycans more (Figure 4).
In summary, as predicted by the structural modeling of the glycomics data, the neutralizing capacity of some BnAbs with glycan-(co)dependent epitopes ranging from the base of the trimer to its apex was impacted by N262 glycan. Specifically, PGT151 and 35O22 (gp120/gp41 interface), 2G12 and VRC24 (V3, C3, and V4 regions), and PG16 and PGT145 (trimer apex) exhibited reduced neutralization capacity against Env-pseudotyped viruses with N262 NGS. Together, these results confirm our glycomics and structural modeling data that suggest that the local N262 glycan has a ripple effect on the Env glycan shield structure and the Env conformation with functional implications for virion infectivity.
Discussion
Results in this study provide new insights into how changes in glycosylation sites of the HIV-1 Env, such as loss of N262, can have ripple effect on the glycan shield with profound implications for Env structure and function. How a mutation in a particular glycan or set of glycans influences the entire glycan shield is difficult to predict. The loss of key glycans may produce a hole in the shield that renders it more vulnerable to neutralization by BnAbs, but where that hole emerges is also difficult to predict. Our results confirm a cluster of glycans proximal to N262 that we and others have previously proposed (Hargett et al., 2019; Lemmin et al., 2017; Seabright et al., 2020). Our analyzed Env variants and mutants represent nearly half of all NGS combinations in the sequences deposited in the LANL HIV database for this N262-cluster of glycans and can serve as a model system to study how glycans affect Env structure and function. High-resolution quantitative glycomics analysis revealed that the N262 NGS affected composition of several glycans at distant locations, by altering their processing, showing how N262 and other glycans in the proximity work together. Molecular modeling and MDS revealed that some of the affected glycans are spatially close to N262, in the so-called high-mannose patch, and showed that some of these glycans can move to compensate for a loss of a glycan. Other affected glycans were in the apex of the trimer. These site-specific alterations of Env glycans have significant consequences, as they impact recognition by BnAbs and interaction with CD4 receptor. Although our MDS model was limited to Man5 glycans, our neutralization analysis on HIV-1 pseudotyped virions strongly corroborated the models, glycomics, and other molecular data. In our studies that focused on the N262-anchored cluster of glycans within the high-mannose patch, the glycan-glycan interactions are dictated more by the interactions of adjacent glycan cores (stalks) in close proximity rather than the branches of individual glycans. Still, having Man5 glycans at all sights in the MDS likely underestimated the influence of the glycans on the Env structure. Nevertheless, using the Man5 model should provide a reasonable first approximation of the glycan shielding and movements.
By using Env with a naturally occurring mutation at the N262 NGS (WEAU-d16), we have resolved a previously encountered problem, wherein an artificial removal of N262 NGS led to Env lysosomal degradation and, thus, production of noninfectious virions (Mathys et al., 2014; Wang et al., 2013). HIV-1 Env with naturally absent N262 NGS are rare; analysis of a database of 6,223 HIV-1 Env sequences revealed that N262 potential NGS was in 99.65% of all sequences (Hargett et al., 2019). In this study, we used Env from an early virus (WEAU-d16) naturally without N262 NGS and introduced the N262 NGS abrogating mutation into the matching chronic-stage virus (WEAU-d391) to generate a variant without N262 NGS (WEAU-d391-S264N). For comparison, we also engineered a WEAU-d16 variant (WEAU-d16-N264S) with added N262 NGS. Our results provide strong evidence that the NGS in proximity to N262 work as a functional HMP-microdomain composed of N262, N448, N301, N332, N295, and N413a. When all members of this cluster of glycans anchored by the glycan at N262 are not present in a given variant (including N262 itself), other glycans can shift to fulfill alternate functional roles. However, there is often an immune evasion consequence to be considered. As we have previously proposed, there are optimal combinations in this glycan microdomain (Hargett et al., 2019). There are also suboptimal NGS combinations and densities that allow a given variant to survive until a better combination arises (or returns) due to mutation over time. Importantly, our data suggest that this HMP-microdomain has structural and functional roles that must be balanced with their role in immune evasion.
The first step in HIV-1 host-cell entry is the interaction between Env and CD4 receptor (Maddon et al., 1986; McDougal et al., 1986). Our experiments showed that the N262 glycan influenced Env-CD4 receptor interaction, further extending previous studies (Mathys et al., 2014; Wang et al., 2013). Specifically, Env-pseudotyped viruses with the N262 glycan (WEAU-d391, WEAU-d16-N264S) had higher infectivity and were more sensitive to inhibition of viral entry by sCD4 than variants without N262 NGS (WEAU-d16, WEAU-d391-S264N). Recombinant trimeric gp120 was affected in the same manner: the variants with N262 glycan exhibited higher binding to CD4 compared with the variants without N262 NGS. As N262 NGS is not a part of CD4-binding site, we speculated that the observed differences in CD4 binding represent a ripple effect of N262 glycan. Possible mechanisms may include (1) destabilization of the Env trimer due to the loss of glycan and side-chain interactions with amino acid residues within β22 strand, demonstrated by crystallographic and electron microscopy data (Kong et al., 2015; Ozorowski et al., 2017); (2) an increase in Env trimer flexibility through fewer glycan-glycan interactions within the N-glycan shield, manifested by a lower betweenness centrality within protomers and an increase in processed N-glycans; and (3) partial occlusion by N-glycans spatially close to the CD4-binding site, such as N276, which has relatively more HexNAc4Hex5Fuc1 glycan than HexNAc4Hex3Fuc1 glycan, as shown for WEAU-d16 versus WEAU-d391 Env. Furthermore, the discrepancy between results for virus-entry inhibition by sCD4 (Figure 4A) and virus neutralization by CD4-binding site-specific BnAbs VRC01 and b12 (Table 1) illustrates the complexity of interactions of HIV-1 Env with different reagents, such as sCD4 versus different BnAbs specific for CD4-binding site (Cheng et al., 2018).
A published structure of fully glycosylated HIV-1 gp120 core revealed the N262 glycan to be buried within a weak negatively charged protein cleft bridging the inner and outer domains of gp120 (Kong et al., 2015). Upon CD4 binding, the D1 arm of the N262 glycan moves by approximately 5Å into a new pocket, stabilizing an alpha helix in the CD4-bound state (Kong et al., 2015; Ozorowski et al., 2017). A loss of the N262 glycan may destabilize the gp120 core, thus affecting the affinity of CD4-binding pocket. In fact, our COM calculations from the MDS of WEAU Env trimer variants have suggested that, in the absence of N262, other oligosaccharides, such as N448, shift toward this cleft, partially complementing the role of N262 glycan.
Our glycomics analyses revealed an overall increase in processed oligosaccharides in gp120 trimers without N262, especially at NGS in the vicinity of N262 (Hargett et al., 2019). Even though the CD4-binding pocket is highly conserved and only peripherally surrounded by oligosaccharides (Kwong et al., 1998), structural studies reveal an impact of glycans, such as N276, spatially close to the CD4-binding pocket (Ozorowski et al., 2017; Stewart-Jones et al., 2016). In fact, MDS of the BG505 SOSIP Env trimer suggested that only one CD4-binding pocket is accessible at a time, due to oligosaccharides occluding the pocket on other protomers (Lemmin et al., 2017). Notably, WEAU-d391, the Env that yields the most infective virions among all tested WEAU variants, had the least processed N276 N-glycans. Thus, we can hypothesize that the extent of glycan processing at NGS spatially close to the CD4-binding pocket can influence accessibility of CD4-binding site. This conclusion is consistent with results of a recent MDS study that showed that in the absence of N301 glycan, another glycan that shields V3 loop and CD4-binding-site epitopes, several other glycans can structurally rearrange to maintain the glycan shield (Ferreira et al., 2018). Based on our recent analysis of combinations of glycans in different regions of Env trimer, we propose that some glycan microdomains would only have a limited number of NGS combinations that maintain the glycan shield (Hargett et al., 2019). The functional data and MDS analyses in this study corroborate this idea and further imply that variations in the density of glycans within glycan-cluster unit can alter Env function. Importantly, these aspects can now be effectively characterized by tracking differences in N-glycan heterogeneity profiles with site-specific resolution across Env variants, as demonstrated here.
Furthermore, our data have also revealed that the N262 NGS impacts neutralization sensitivity to some BnAbs. We observed better neutralization by PGT151, 35O22, VRC24, 2G12, VRC24, PG9, PG16, and PGT145 BnAbs for virions pseudotyped with Env without N262. Notably, the absence of the N262 glycan had little effect on BnAbs PGT121 and PGT122 that all recognize an epitope within the high-mannose patch that involves N332 NGS (Table 1). These observations are consistent with the glycomics profiling (Figure 6) and MDS COM calculations (Figure 8), which have shown that N332 has a similar glycan profile and COM location in all WEAU variants. This would imply that N332 glycan is less structurally/functionally dependent on the cluster of glycans surrounding N262 glycans. Conversely, N413 glycan that originates further away from N262 than N332, does appear to play a significant role in maintaining the glycan structural density for this N262-based cluster of glycans based on our glycomic, molecular, and MDS data, and the published structure of the clade G X1193.c1 glycosylated structure (Stewart-Jones et al., 2016). The Env glycan shield is a complex network of glycan-glycan interactions (Kwong et al., 1998; Lemmin et al., 2017; Li et al., 1993; Pollakis et al., 2001; Stewart-Jones et al., 2016). We are only now beginning to understand how to define and probe how those clusters of glycans work together to maintain function and structure of the Env in the context of constant mutation process (Hargett et al., 2019). Rather than single glycan frequencies, regions of HIV Env glycans need to be defined and tested as working-unit clusters.
In summary, this study revealed how a naturally occurring N-glycan mutation can affect function of HIV-1 Env through altered glycosylation. Although there are many other Env variants impacting N-glycan shield to be explored, this study provides new insight into complex effects of Env glycans on HIV biology. By utilizing molecular experiments, structural analysis, and glycomics profiling, we can begin to understand the limitations of the HIV-1 “balancing act” between the viral immune escape and infectivity.
Limitations of the Study
In this study, we used two naturally occurring variants of Env gp120 derived from WEAU-d16 and WEAU-d391 viruses from a single infected individual. These gp120 variants were selected based on their unique characteristics (Wei et al., 2003; Keele et al., 2008). It is to be noted that the pseudotyped viruses and recombinant gp120 trimers in this study were produced in 293F cells and not in the HIV-1 natural host cells, T cells, or monocytes/macrophages. 293F cells, derived from HEK293 adherent cells, have been adapted to a growth in suspension in serum-free medium and have been used by us as well as by other investigators. We used 293F, rather than the natural host cells, as the system provides multiple advantages, including high-level glycoprotein expression and avoiding problems with serum use in cell-culture medium. However, we and others have shown that Env glycosylation varies depending on the producing cells (Cao et al., 2018; Go et al., 2013, 2014, 2015; Raska et al., 2010, 2014; Shen et al., 2014). For example, T cell lines and muscle cells produce gp120 trimers with elevated representation of hybrid glycans. Furthermore, glycosylation of soluble trimers, such as SOSIP, can differ from glycosylation of trimeric Env on the virions (Cao et al., 2018). In spite of these limitations, this study demonstrates how changes in individual glycan units can alter molecular dynamics and processing of the HIV-1 Env-glycan shield and, consequently, Env function. These findings together provide additional information relevant for the enveloped virus structural biology as well as the ongoing development of HIV-1 vaccines based on the viral spike.
Resource Availability
Lead Contact
Requests for materials and communications with the journal should be addressed to J.N. (jannovak@uab.edu).
Materials Availability
All unique/stable reagents generated in this study are available via the Lead Contact with a completed Materials Transfer Agreement.
Data and Code Availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [1; http://www.ebi.ac.uk/pride] partner repository with the dataset identifier PXD017941.
[1] Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, Pérez E, Uszkoreit J, Pfeuffer J, Sachsenberg T, Yilmaz S, Tiwary S, Cox J, Audain E, Walzer M, Jarnuczak AF, Ternent T, Brazma A, Vizcaíno JA (2019). The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 47(D1):D442-D450 (PubMed ID: 30395289).
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
Support for this work was provided by the National Institutes of Health (NIH) grant GM098539, by the Intramural Research Program of the Vaccine Research Center, NIAID, NIH, and by NIH T32 fellowship GM008111. This work was also supported in part by the University of Alabama at Birmingham (UAB) Center for AIDS Research (AI027767) developmental grant and a pilot grant from UAB School of Medicine. MR was supported in part by the UP grant IGA_LF_2020_016 and a grant of Ministry of School and Education, Youth and Sport of the Czech Republic, CZ.02.1.01/0.0/0.0/16_025/0007397. BK was supported with Federal funds from the National Cancer Institute, NIH, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. The authors would like to thank Beatrice Hahn, George Shaw, and Jiri Mestecky for helpful discussions.
Author Contributions
Z.M., M.R., M.B.R., and J.N. designed the study. B.F.K., M.R., and J.N. selected WEAU gp120 variants from early and late stages of HIV-1 infection. Q.W., A.A.H., B.K., A.D., S.H., and R.B. carried out the research. S.L.H.and M.S.S. provided materials. R.R. prepared, carried out, and analyzed the MDS. C.H.S performed network analysis of MDS trajectories. S.K.F. assisted in analyzing glycan flexibility in MDS trajectories. O.K., G.Y.C., P.D.K., Z.M., M.R., M.B.R., and J.N. supervised the research. Q.W., A.A.H., Z.M., M.R., M.B.R., and J.N. drafted the manuscript. Q.W., A.A.H., B.K., R.R., B.F.K., O.K., P.D.K., Z.M., M.R., M.B.R., and J.N. edited the manuscript. All authors analyzed data and commented on the drafts.
Declaration of Interests
The authors declare no competing interests.
Published: November 20, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.101711.
Contributor Information
Matthew B. Renfrow, Email: renfrow@uab.edu.
Jan Novak, Email: jannovak@uab.edu.
Supplemental Information
References
- Abrahams M.R., Anderson J.A., Giorgi E.E., Seoighe C., Mlisana K., Ping L.H., Athreya G.S., Treurnicht F.K., Keele B.F., Wood N. Quantitating the multiplicity of infection with human immunodeficiency virus type 1 subtype C reveals a non-Poisson distribution of transmitted variants. J. Virol. 2009;83:3556–3567. doi: 10.1128/JVI.02132-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens A.J., Harvey D.J., Milne E., Cupo A., Kumar A., Zitzmann N., Struwe W.B., Moore J.P., Crispin M. Molecular architecture of the cleavage-dependent mannose patch on a soluble HIV-1 envelope glycoprotein trimer. J. Virol. 2017;91 doi: 10.1128/JVI.01894-16. e01894–01816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner C., Lee J.H., Sliepen K., Derking R., Falkowska E., de la Pena A.T., Cupo A., Julien J.P., van Gils M., Lee P.S. Structural delineation of a quaternary, cleavage-dependent epitope at the gp41-gp120 interface on intact HIV-1 Env trimers. Immunity. 2014;40:669–680. doi: 10.1016/j.immuni.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonsignori M., Montefiori D.C., Wu X., Chen X., Hwang K.K., Tsao C.Y., Kozink D.M., Parks R.J., Tomaras G.D., Crump J.A. Two distinct broadly neutralizing antibody specificities of different clonal lineages in a single HIV-1-infected donor: implications for vaccine design. J. Virol. 2012;86:4688–4692. doi: 10.1128/JVI.07163-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L., Pauthner M., Andrabi R., Rantalainen K., Berndsen Z., Diedrich J.K., Menis S., Sok D., Bastidas R., Park S.R. Differential processing of HIV envelope glycans on the virus and soluble recombinant trimer. Nat. Commun. 2018;9:3693. doi: 10.1038/s41467-018-06121-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H.D., Grimm S.K., Gilman M.S., Gwom L.C., Sok D., Sundling C., Donofrio G., Karlsson Hedestam G.B., Bonsignori M., Haynes B.F. Fine epitope signature of antibody neutralization breadth at the HIV-1 envelope CD4-binding site. JCI Insight. 2018;3:e97018. doi: 10.1172/jci.insight.97018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun H.M., Carpenter R.J., Macalino G.E., Crum-Cianflone N.F. The role of sexually transmitted infections in HIV-1 progression: a comprehensive review of the literature. J. Sex. Transm. Dis. 2013;2013:176459. doi: 10.1155/2013/176459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis K.L., Gray E.S., Moore P.L., Decker J.M., Salomon A., Montefiori D.C., Graham B.S., Keefer M.C., Pinter A., Morris L. High titer HIV-1 V3-specific antibodies with broad reactivity but low neutralizing potency in acute infection and following vaccination. Virology. 2009;387:414–426. doi: 10.1016/j.virol.2009.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doores K.J., Kong L., Krumm S.A., Le K.M., Sok D., Laserson U., Garces F., Poignard P., Wilson I.A., Burton D.R. Two classes of broadly neutralizing antibodies within a single lineage directed to the high-mannose patch of HIV envelope. J. Virol. 2015;89:1105–1118. doi: 10.1128/JVI.02905-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euler Z., Bunnik E.M., Burger J.A., Boeser-Nunnink B.D., Grijsen M.L., Prins J.M., Schuitemaker H. Activity of broadly neutralizing antibodies, including PG9, PG16, and VRC01, against recently transmitted subtype B HIV-1 variants from early and late in the epidemic. J. Virol. 2011;85:7236–7245. doi: 10.1128/JVI.00196-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkowska E., Le K.M., Ramos A., Doores K.J., Lee J.H., Blattner C., Ramirez A., Derking R., van Gils M.J., Liang C.H. Broadly neutralizing HIV antibodies define a glycan-dependent epitope on the prefusion conformation of gp41 on cleaved envelope trimers. Immunity. 2014;40:657–668. doi: 10.1016/j.immuni.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira R., Grant O., Moyo T., Dorfman J., Woods R., Travers S., Wood N. Structural rearrangements maintain the glycan shield of an HIV-1 envelope trimer after the loss of a glycan. Sci. Rep. 2018;8:15031. doi: 10.1038/s41598-018-33390-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer W., Ganusov V.V., Giorgi E.E., Hraber P.T., Keele B.F., Leitner T., Han C.S., Gleasner C.D., Green L., Lo C.C. Transmission of single HIV-1 genomes and dynamics of early immune escape revealed by ultra-deep sequencing. PLoS One. 2010;5:e12303. doi: 10.1371/journal.pone.0012303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiev I.S., Doria-Rose N.A., Zhou T., Kwon Y.D., Staupe R.P., Moquin S., Chuang G.Y., Louder M.K., Schmidt S.D., Altae-Tran H.R. Delineating antibody recognition in polyclonal sera from patterns of HIV-1 isolate neutralization. Science. 2013;340:751–756. doi: 10.1126/science.1233989. [DOI] [PubMed] [Google Scholar]
- Go E.P., Ding H., Zhang S., Ringe R.P., Nicely N., Hua D., Steinbock R.T., Golabek M., Alin J., Alam S.M. Glycosylation benchmark profile for HIV-1 envelope glycoprotein production based on eleven Env trimers. J. Virol. 2017;91 doi: 10.1128/JVI.02428-16. e02428–02416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go E.P., Herschhorn A., Gu C., Castillo-Menendez L., Zhang S., Mao Y., Chen H., Ding H., Wakefield J.K., Hua D. Comparative analysis of the glycosylation profiles of membrane-anchored HIV-1 envelope glycoprotein trimers and soluble gp140. J. Virol. 2015;89:8245–8257. doi: 10.1128/JVI.00628-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go E.P., Hua D., Desaire H. Glycosylation and disulfide bond analysis of transiently and stably expressed clade C HIV-1 gp140 trimers in 293T cells identifies disulfide heterogeneity present in both proteins and differences in O-linked glycosylation. J. Proteome Res. 2014;13:4012–4027. doi: 10.1021/pr5003643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go E.P., Liao H.X., Alam S.M., Hua D., Haynes B.F., Desaire H. Characterization of host-cell line specific glycosylation profiles of early transmitted/founder HIV-1 gp120 envelope proteins. J. Proteome Res. 2013;12:1223–1234. doi: 10.1021/pr300870t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargett A.A., Wei Q., Knoppova B., Hall S., Huang Z.Q., Prakash A., Green T.J., Moldoveanu Z., Raska M., Novak J. Defining HIV-1 Envelope N-glycan microdomains through site-specific heterogeneity profiles. J. Virol. 2019;93 doi: 10.1128/JVI.01177-18. e01177–01118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessell A.J., Rakasz E.G., Poignard P., Hangartner L., Landucci G., Forthal D.N., Koff W.C., Watkins D.I., Burton D.R. Broadly neutralizing human anti-HIV antibody 2G12 is effective in protection against mucosal SHIV challenge even at low serum neutralizing titers. PLoS Pathog. 2009;5:e1000433. doi: 10.1371/journal.ppat.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Kang B.H., Pancera M., Lee J.H., Tong T., Feng Y., Imamichi H., Georgiev I.S., Chuang G.Y., Druz A. Broad and potent HIV-1 neutralization by a human antibody that binds the gp41-gp120 interface. Nature. 2014;515:138–142. doi: 10.1038/nature13601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J., Whitford W., Wagner F., Kutsch O. Optimization of HIV-1 infectivity assays. Biotechniques. 2007;43:589–590. doi: 10.2144/000112624. [DOI] [PubMed] [Google Scholar]
- Keele B.F., Giorgi E.E., Salazar-Gonzalez J.F., Decker J.M., Pham K.T., Salazar M.G., Sun C., Grayson T., Wang S., Li H. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. U S A. 2008;105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L., Wilson I.A., Kwong P.D. Crystal structure of a fully glycosylated HIV-1 gp120 core reveals a stabilizing role for the glycan at Asn262. Proteins. 2015;83:590–596. doi: 10.1002/prot.24747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumm S.A., Mohammed H., Le K.M., Crispin M., Wrin T., Poignard P., Burton D.R., Doores K.J. Mechanisms of escape from the PGT128 family of anti-HIV broadly neutralizing antibodies. Retrovirology. 2016;13:8. doi: 10.1186/s12977-016-0241-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong P.D., Wyatt R., Robinson J., Sweet R.W., Sodroski J., Hendrickson W.A. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Leaman D.P., Kim A.S., Torrents de la Pena A., Sliepen K., Yasmeen A., Derking R., Ramos A., de Taeye S.W., Ozorowski G. Antibodies to a conformational epitope on gp41 neutralize HIV-1 by destabilizing the Env spike. Nat. Commun. 2015;6:8167. doi: 10.1038/ncomms9167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Ozorowski G., Ward A.B. Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science. 2016;351:1043–1048. doi: 10.1126/science.aad2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W.R., Syu W.J., Du B., Matsuda M., Tan S., Wolf A., Essex M., Lee T.H. Nonrandom distribution of gp120 N-linked glycosylation sites important for infectivity of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. U S A. 1992;89:2213–2217. doi: 10.1073/pnas.89.6.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmin T., Soto C., Stuckey J., Kwong P.D. Microsecond dynamics and network analysis of the HIV-1 SOSIP Env trimer reveal collective behavior and conserved microdomains of the glycan shield. Structure. 2017;25:1631–1639 e1632. doi: 10.1016/j.str.2017.07.018. [DOI] [PubMed] [Google Scholar]
- Li Y., Luo L., Rasool N., Kang C.Y. Glycosylation is necessary for the correct folding of human immunodeficiency virus gp120 in CD4 binding. J. Virol. 1993;67:584–588. doi: 10.1128/jvi.67.1.584-588.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H.X., Tsao C.Y., Alam S.M., Muldoon M., Vandergrift N., Ma B.J., Lu X., Sutherland L.L., Scearce R.M., Bowman C. Antigenicity and immunogenicity of transmitted/founder, consensus, and chronic envelope glycoproteins of human immunodeficiency virus type 1. J. Virol. 2013;87:4185–4201. doi: 10.1128/JVI.02297-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Bartesaghi A., Borgnia M.J., Sapiro G., Subramaniam S. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455:109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddon P.J., Dalgleish A.G., McDougal J.S., Clapham P.R., Weiss R.A., Axel R. The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell. 1986;47:333–348. doi: 10.1016/0092-8674(86)90590-8. [DOI] [PubMed] [Google Scholar]
- Mathys L., Francois K.O., Quandte M., Braakman I., Balzarini J. Deletion of the highly conserved N-glycan at Asn260 of HIV-1 gp120 affects folding and lysosomal degradation of gp120, and results in loss of viral infectivity. PLoS One. 2014;9:e101181. doi: 10.1371/journal.pone.0101181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCurley N.P., Domi A., Basu R., Saunders K.O., LaBranche C.C., Montefiori D.C., Haynes B.F., Robinson H.L. HIV transmitted/founder vaccines elicit autologous tier 2 neutralizing antibodies for the CD4 binding site. PLoS One. 2017;12:e0177863. doi: 10.1371/journal.pone.0177863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDougal J.S., Kennedy M.S., Sligh J.M., Cort S.P., Mawle A., Nicholson J.K. Binding of HTLV-III/LAV to T4+ T cells by a complex of the 110K viral protein and the T4 molecule. Science. 1986;231:382–385. doi: 10.1126/science.3001934. [DOI] [PubMed] [Google Scholar]
- McLellan J.S., Pancera M., Carrico C., Gorman J., Julien J.P., Khayat R., Louder R., Pejchal R., Sastry M., Dai K. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montefiori D.C. Measuring HIV neutralization in a luciferase reporter gene assay. Methods Mol. Biol. 2009;485:395–405. doi: 10.1007/978-1-59745-170-3_26. [DOI] [PubMed] [Google Scholar]
- Murin C.D., Julien J.P., Sok D., Stanfield R.L., Khayat R., Cupo A., Moore J.P., Burton D.R., Wilson I.A., Ward A.B. Structure of 2G12 Fab2 in complex with soluble and fully glycosylated HIV-1 Env by negative-stain single-particle electron microscopy. J. Virol. 2014;88:10177–10188. doi: 10.1128/JVI.01229-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozorowski G., Pallesen J., de Val N., Lyumkis D., Cottrell C.A., Torres J.L., Copps J., Stanfield R.L., Cupo A., Pugach P. Open and closed structures reveal allostery and pliability in the HIV-1 envelope spike. Nature. 2017;547:360–363. doi: 10.1038/nature23010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pejchal R., Doores K.J., Walker L.M., Khayat R., Huang P.S., Wang S.K., Stanfield R.L., Julien J.P., Ramos A., Crispin M. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science. 2011;334:1097–1103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollakis G., Kang S., Kliphuis A., Chalaby M.I., Goudsmit J., Paxton W.A. N-linked glycosylation of the HIV type-1 gp120 envelope glycoprotein as a major determinant of CCR5 and CXCR4 coreceptor utilization. J. Biol. Chem. 2001;276:13433–13441. doi: 10.1074/jbc.M009779200. [DOI] [PubMed] [Google Scholar]
- Pritchard L.K., Spencer D.I., Royle L., Bonomelli C., Seabright G.E., Behrens A.J., Kulp D.W., Menis S., Krumm S.A., Dunlop D.C. Glycan clustering stabilizes the mannose patch of HIV-1 and preserves vulnerability to broadly neutralizing antibodies. Nat. Commun. 2015;6:7479. doi: 10.1038/ncomms8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raska M., Czernekova L., Moldoveanu Z., Zachova K., Elliott M.C., Novak Z., Hall S., Hoelscher M., Maboko L., Brown R. Differential glycosylation of envelope gp120 is associated with differential recognition of HIV-1 by virus-specific antibodies and cell infection. AIDS Res. Ther. 2014;11:23. doi: 10.1186/1742-6405-11-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raska M., Moldoveanu Z., Novak J., Hel Z., Novak L., Bozja J., Compans R.W., Yang C., Mestecky J. Delivery of DNA HIV-1 vaccine to the liver induces high and long-lasting humoral immune responses. Vaccine. 2008;26:1541–1551. doi: 10.1016/j.vaccine.2008.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raska M., Takahashi K., Czernekova L., Zachova K., Hall S., Moldoveanu Z., Elliott M.C., Wilson L., Brown R., Jancova D. Glycosylation patterns of HIV-1 gp120 depend on the type of expressing cells and affect antibody recognition. J. Biol. Chem. 2010;285:20860–20869. doi: 10.1074/jbc.M109.085472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman D.D., Wrin T., Little S.J., Petropoulos C.J. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. U S A. 2003;100:4144–4149. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzuto C.D., Wyatt R., Hernandez-Ramos N., Sun Y., Kwong P.D., Hendrickson W.A., Sodroski J. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science. 1998;280:1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- Seabright G.E., Cottrell C.A., van Gils M.J., D'Addabbo A., Harvey D.J., Behrens A.J., Allen J.D., Watanabe Y., Scaringi N., Polveroni T.M. Networks of HIV-1 envelope glycans maintain antibody epitopes in the face of glycan additions and deletions. Structure. 2020;28:897–909. doi: 10.1016/j.str.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen R., Raska M., Bimczok D., Novak J., Smith P.D. HIV-1 envelope glycan moieties modulate HIV-1 transmission. J. Virol. 2014;88:14258–14267. doi: 10.1128/JVI.02164-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivatare V.S., Shivatare S.S., Lee C.D., Liang C.H., Liao K.S., Cheng Y.Y., Saidachary G., Wu C.Y., Lin N.H., Kwong P.D. Unprecedented role of hybrid N-glycans as ligands for HIV-1 broadly neutralizing antibodies. J. Am. Chem. Soc. 2018;140:5202–5210. doi: 10.1021/jacs.8b00896. [DOI] [PubMed] [Google Scholar]
- Stewart-Jones G.B., Soto C., Lemmin T., Chuang G.Y., Druz A., Kong R., Thomas P.V., Wagh K., Zhou T., Behrens A.J. Trimeric HIV-1-Env structures define glycan shields from clades A, B, and G. Cell. 2016;165:813–826. doi: 10.1016/j.cell.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanstrom R., Coffin J. HIV-1 pathogenesis: the virus. Cold Spring Harb. Perspect. Med. 2012;2:a007443. doi: 10.1101/cshperspect.a007443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaras G.D., Yates N.L., Liu P., Qin L., Fouda G.G., Chavez L.L., Decamp A.C., Parks R.J., Ashley V.C., Lucas J.T. Initial B-cell responses to transmitted human immunodeficiency virus type 1: virion-binding immunoglobulin M (IgM) and IgG antibodies followed by plasma anti-gp41 antibodies with ineffective control of initial viremia. J. Virol. 2008;82:12449–12463. doi: 10.1128/JVI.01708-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Nie J., Prochnow C., Truong C., Jia Z., Wang S., Chen X.S., Wang Y. A systematic study of the N-glycosylation sites of HIV-1 envelope protein on infectivity and antibody-mediated neutralization. Retrovirology. 2013;10:14. doi: 10.1186/1742-4690-10-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X., Decker J.M., Wang S., Hui H., Kappes J.C., Wu X., Salazar-Gonzalez J.F., Salazar M.G., Kilby J.M., Saag M.S. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- Wyatt R., Kwong P.D., Desjardins E., Sweet R.W., Robinson J., Hendrickson W.A., Sodroski J.G. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393:705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [1; http://www.ebi.ac.uk/pride] partner repository with the dataset identifier PXD017941.
[1] Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, Pérez E, Uszkoreit J, Pfeuffer J, Sachsenberg T, Yilmaz S, Tiwary S, Cox J, Audain E, Walzer M, Jarnuczak AF, Ternent T, Brazma A, Vizcaíno JA (2019). The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 47(D1):D442-D450 (PubMed ID: 30395289).