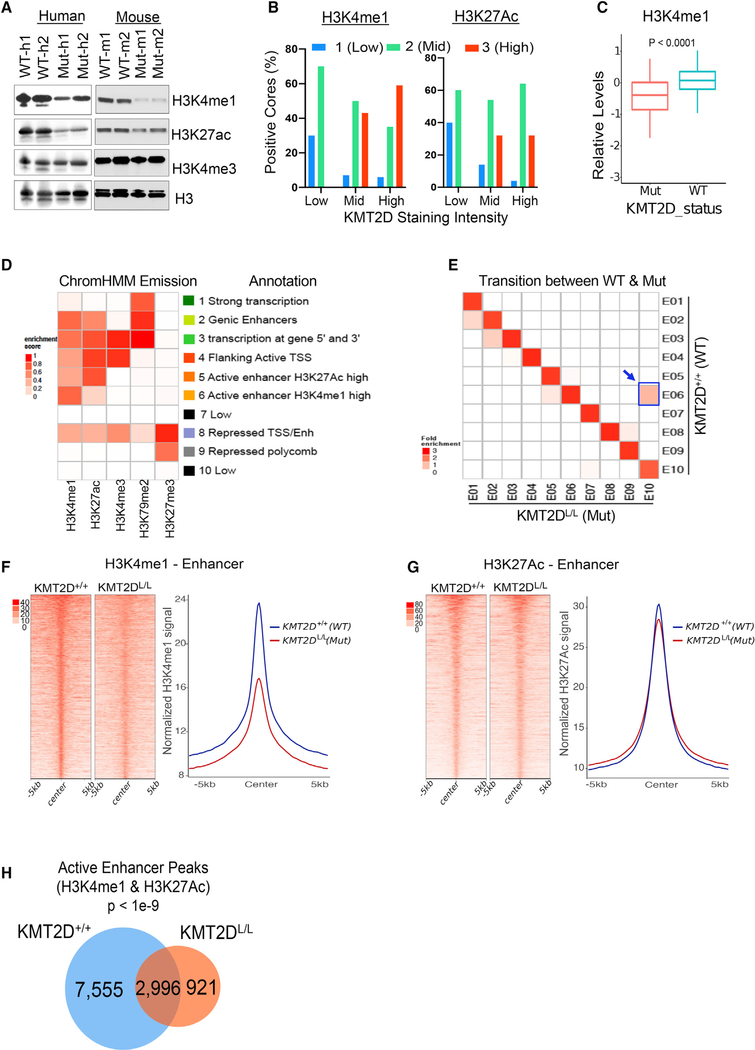
Figure 5. KMT2D Loss Is Associated with Loss of H3K4me1-Marked Enhancers.
(A) Western blot showing total H3K4me1, H3K27ac, H3K4me3, and H3 in KMT2D mutant (Mut-m1, Mut-m2, Mut-h1, and Mut-h2) and WT (WT-m1, WT-m2, WT-h1, and WT-h2) murine (right) and human (left) melanoma cells.
(B) Bar chart showing immunohistochemistry-based staining of melanoma tumors (MDACC TCGA samples) in a tissue microarray for KMT2D, H3K4me1 (left panel), and H3K27ac (right panel). KMT2D intensity was grouped into low (score of 1 or 1.5, n = 10), mid (score of 2, n = 57), and high (score of 3, n = 33) groups, and percent positive cores with similar intensity groups for H3K4me1 or H3K27ac were plotted on y axis.
(C) Boxplot showing relative levels of H3K4me1 in KMT2D mutant (n = 126) or WT (n = 293) cells using the mass spectrometry data from the CCLE database. The bottom and the top rectangles indicate the first quartile (Q1) and third quartile (Q3), respectively. The horizontal lines in the middle signify the median (Q2), and the vertical lines that extend from the top and the bottom of the plot indicate the maximum and minimum values, respectively.
(D) Emission probabilities of the 10-state ChromHMM model on the basis of ChIP-seq profiles of five histone marks (shown in x axis). Each row represents one chromatin state, and each column corresponds to one chromatin mark. The intensity of the color in each cell reflects the frequency of occurrence of that mark in the corresponding chromatin state on the scale from 0 (white) to 1 (red). States were manually grouped and given candidate annotations.
(E) Heatmap showing the fold enrichment of chromatin state transitions between KMT2D mutant (KMT2DL/L) and WT (KMT2D+/+) samples for the 10-state model defined by the ChromHMM. Color intensities represent the relative fold enrichment. Blue box and arrow point to active enhancer state switch.
(F and G) Heatmaps (left panels) and average intensity curves (right panels) of ChIP-seq reads (RPKM, reads per kilobase of transcript, per million mapped reads) for H3K4me1 (F) and H3K27ac (G) at typical enhancer regions. Enhancers are shown in a 10-kb window centered on the middle of the enhancer in iBIP;KMT2D+/+ and iBIP;KMT2DL/L melanoma tumors.
(H) Venn diagram showing unique or shared H3K4me1 and H3K27Ac co-enriched active enhancers sites in iBIP;KMT2D+/+ and iBIP;KMT2DL/L melanoma tumors.