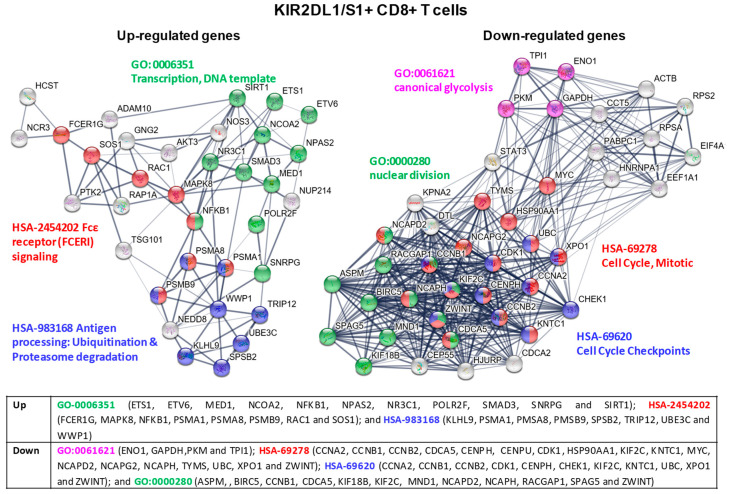
Figure 7.
KIR2DL1/S1+ CD8+ T cell cluster analysis of the protein-protein interaction (PPI) network with STRING database. Hub nodes from differentially expressed genes were identified using Cytoscape software and then represented using STRING to perform the functional and enrichment analysis of these pathways. The PPI network shows up-expressed genes for modules GO-0006351 Transcription, DNA template (green), HSA-2454202 Fcε receptor (FCERI) signaling (red), HSA-983168 Antigen processing: Ubiquitination& Proteasome degradation (blue); and down-expressed genes for modules GO-0061621 canonical glycolysis (purple), HSA-69278 Cell Cycle, Mitotic (red), HSA-69620 Cell Cycle Checkpoints (blue), and GO:0000280 nuclear division (green). Briefly, the parameters used in STRING were as follows: text mining, experiments, databases, co-expression, neighborhood, gene fusion and co-occurrence were used as active interaction sources. The confidence threshold was set at 0.4. The disconnected transcripts were eliminated.