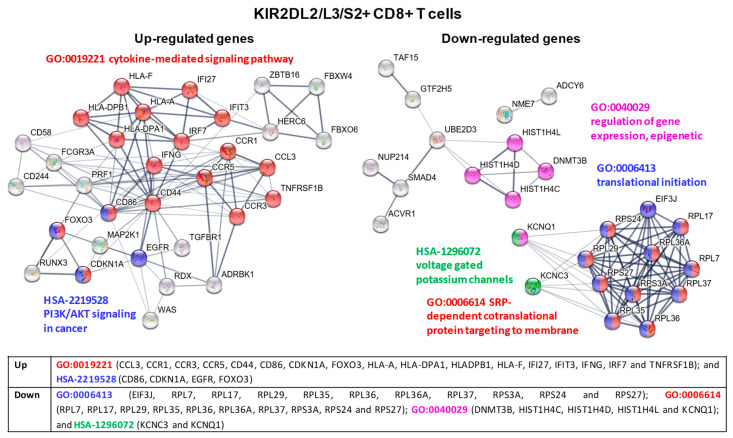
Figure 8.
KIR2DL2/L3/S2+ CD8+ T cell cluster analysis of the protein-protein interaction (PPI) network with STRING database. Hub nodes from differentially expressed genes were identified using Cytoscape software and then represented using STRING to perform the functional and enrichment analysis of these pathways. The PPI network shows up-expressed genes for modules GO-0019221 cytokine-mediated signaling pathway (red) and HSA-2219528 PI3K/AKT Signaling in Cancer (blue); and down-expressed genes for modules GO:0006413 translational initiation (blue), GO-0006614 SRP-dependent cotranslational protein targeting to membrane (red), GO:0040029 regulation of gene expression, epigenetic (purple), and HSA-1296072 voltage gated potassium channels (green). Briefly, the parameters used in STRING were as follows: text mining, experiments, databases, co-expression, neighborhood, gene fusion and co-occurrence were used as active interaction sources. The confidence threshold was set at 0.4. The disconnected transcripts were eliminated.