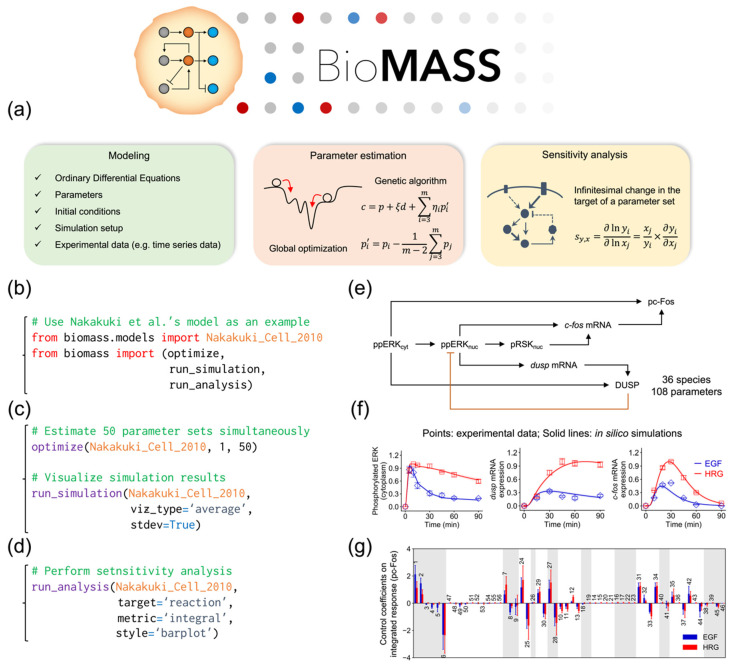
Figure 1.
Overview of the BioMASS workflow. (a) After setting model properties (see Table 1), users can estimate unknown model parameters against experimental observations. Based on optimized parameter sets, users can perform sensitivity analysis to identify critical components or processes for cellular output. This workflow implements the model of Nakakuki et al. (b–d) Example code for model import (b), parameter estimation and visualization of simulation results (c) and sensitivity analysis (d). (e) Core reaction scheme of the Nakakuki model. (f) Simulation results using optimized parameter sets. (g) Output of the sensitivity analysis.