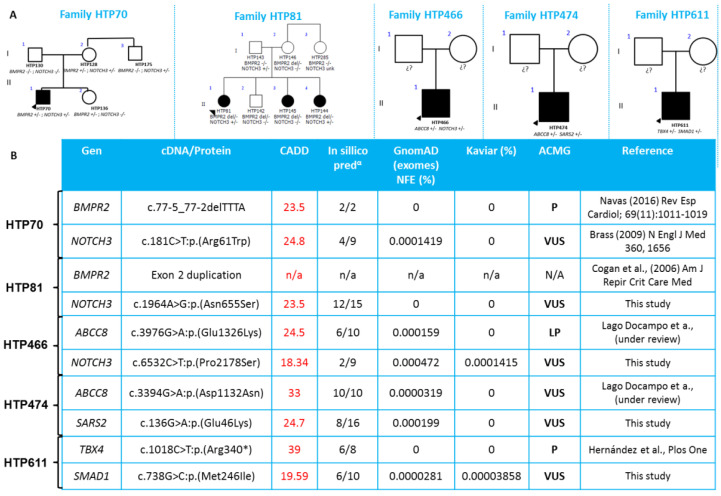
Figure 3.
Pedigree and in silico analysis of the families with more than one variant detected. (A) Genealogy of the families in which segregation analysis was performed (when available). (B) Table of in sillico analysis of the candidate variants, including the ACMG classification as well as the reference in which the variant was described. n/a: not applicable, P: pathogenic, VUS: Variant of unknown significance. α: In silico analysis was performed by dbNSFP counting the ones that predict a deleterious effect over the total available.