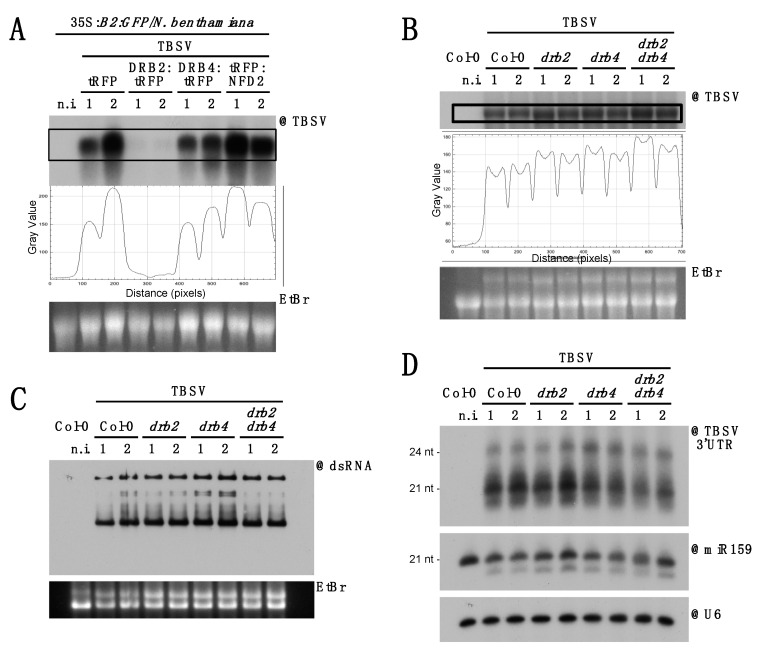
Figure 5.
Involvement of DRB2, DRB4 and NFD2 in TBSV infection. (A) Northern blot analysis of RNA from 35S:B2:GFP/N. benthamiana leaf disks agroinoculated with the constructs indicated and subsequently infected with TBSV. Each sample is a pool of 50–60 leaf disks taken from several infected leaves on 2–3 plants. (B) Northern blot analysis of high molecular weight RNA to detect TBSV at 9 dpi in systemically infected rosette leaves of A. thaliana mutant lines. Quantification of signal intensity is provided. Each sample is a pool of 4–5 plants. (C) Northwestern blot to detect dsRNA in the samples described in (B). In (A–C) EtBr gel staining was used as a loading control. (D) Northern blot analysis of low molecular weight RNA to detect TBSV-derived siRNA. As loading control, the same membrane was separately hybridized to miR159- and snU6-specific probes. In all the blots a non-infected control (n.i.) was included on the far left. Except for the non-infected control, two samples were analyzed per genotype/transgene (1 and 2 in the labels).