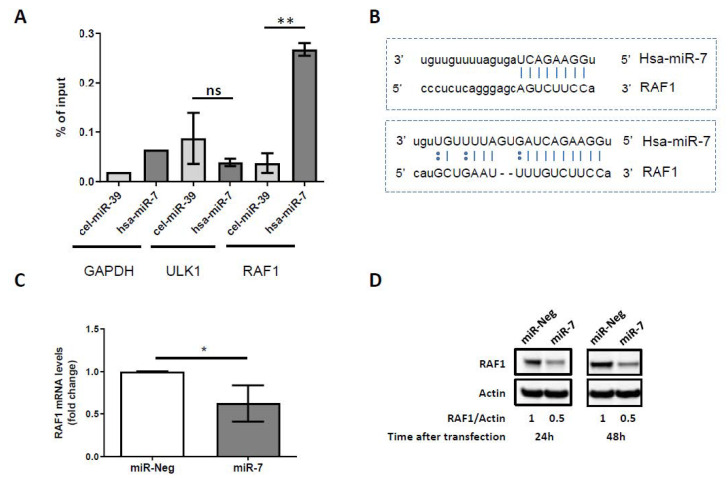
Figure 3.
The identification of RAF1 mRNA as a target of miR-7-5p. (A) The RNAs co-purified with biotinylated miR-7 versus with biotinylated cel-miR-39 (control miRNA from C. elegans) were analyzed by qRT-PCR using PCR primers for two miR-7 predicted target genes (ULK1 and RAF1). RAF1 mRNAs were significantly pulled-down with the biotinylated miR-7 versus cel-miR-39 whereas the ULK1 mRNAs were not. GAPDH mRNA was not predicted as a miR-7 target, and allowed us to evaluate and remove the pulled-down background. Data represent the mean ± SD; n = 3; ns: not significant; ** p ≤ 0.01; unpaired Student’s t test. (B) Sequence alignment between miR-7 and the 3′UTR of RAF1 mRNA identified two complementary sites (www.microrna.org). (C) qPCR analysis of RAF1 mRNA expression in control miRNA (miR-Neg) or miR-7-5p (miR-7) transfected Karpas-299 cells (mean ± SD of independent experiments, n = 4, * p ≤ 0.05; unpaired Student’s t test). (D) Representative western blot showing the RAF1 protein levels 24 or 48 h following control miRNA (miR-Neg) or miR-7-5p (miR-7) transfection in Karpas-299. Actin was used as a loading control. RAF1/Actin densitometric ratios are indicated.