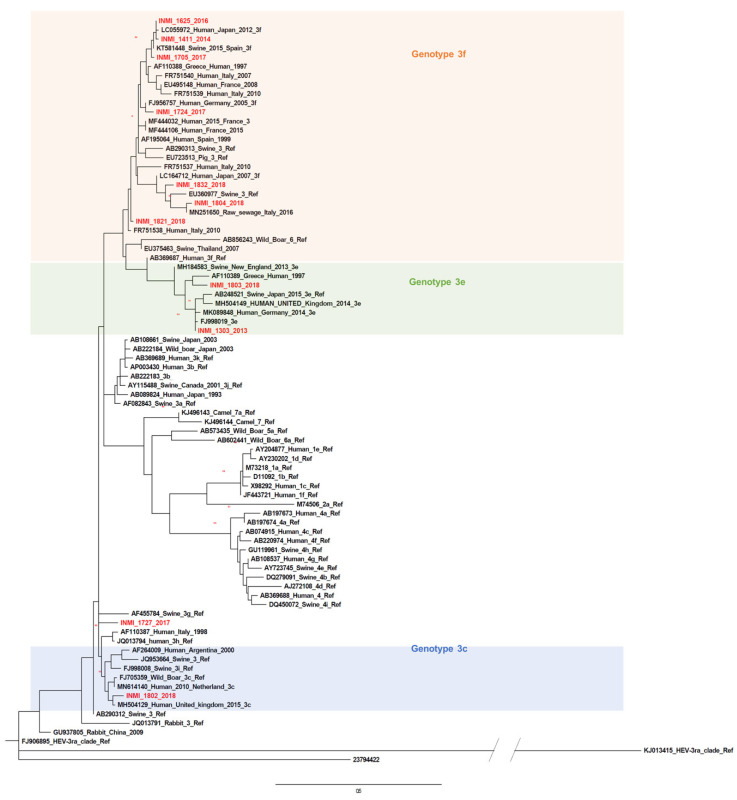
Figure 6.
Phylogenetic analysis based on partial open reading frame 1 (ORF1, 125 nt, nt 78-202) of the HE-JA04-1911 isolate (GenBank accession number AB248521) and patient-derived sequences obtained in this study. The phylogenetic tree includes the prototype strains as indicated by Smith et al. [24] and other sequences isolated from different geographical regions with a high percentage of similarity to our sequences. Previously described HEV GT3 subtypes are indicated. The GT1 sequence was used as the outgroup. The phylogenetic tree was built by using the maximum likelihood method based on the general time reversible model with a bootstrap of 500 replicates. Bootstrap values >60% are indicated on respective branches. The scale bar represents nucleotide substitutions per site.