Extended Data Fig. 1. Detailed overview of CHANGE-seq method.
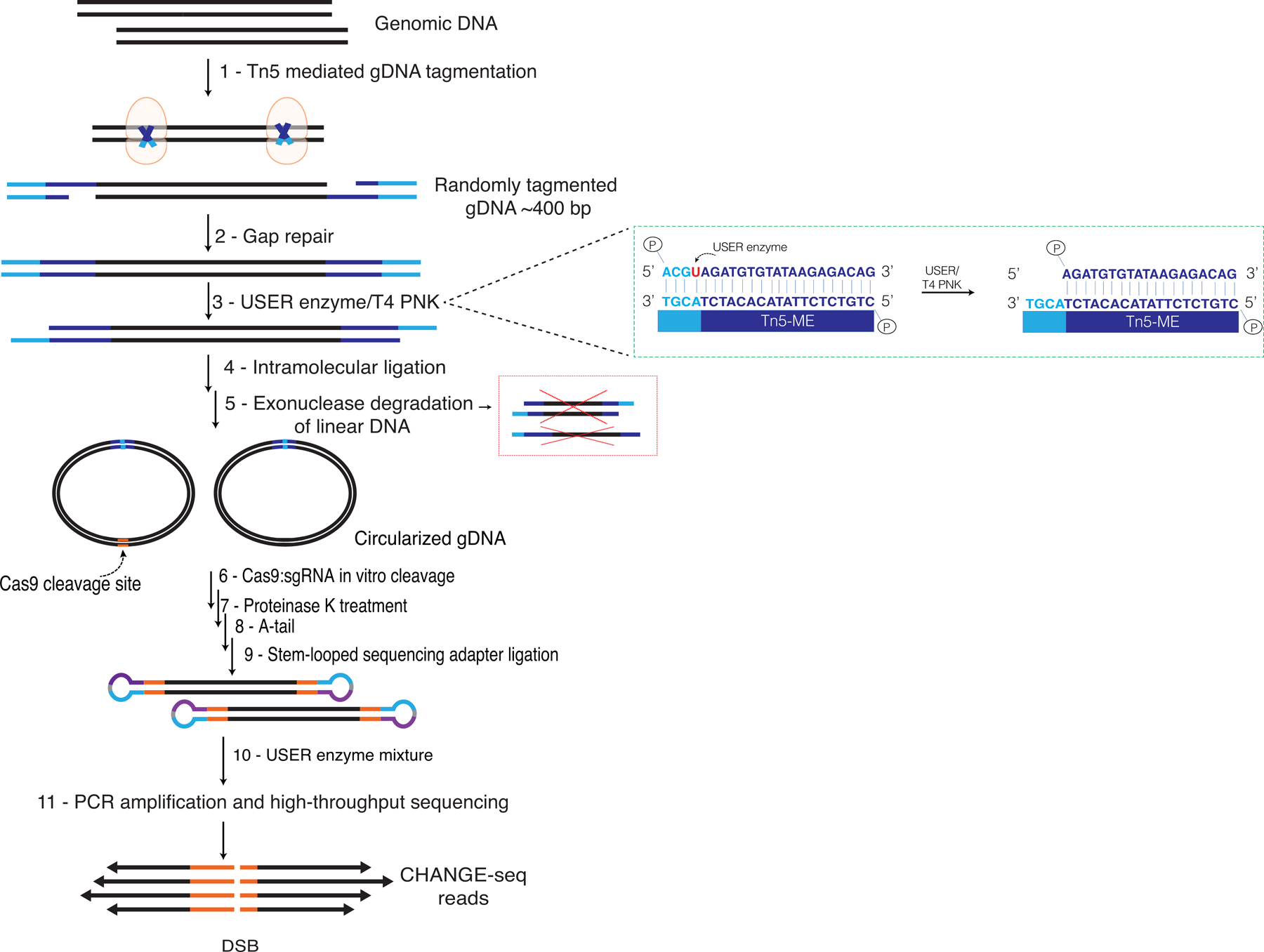
Genomic DNA is randomly tagmented to an average of ~400 bp with a custom Tn5-transposome with an uracil-containing adapter. 9-nt Tn5-generated gaps in the DNA are filled in with a high-fidelity uracil-tolerant U+ polymerase and sealed with Taq DNA ligase. 4 bp overhangs are released with a mixture of USER enzyme and T4 PNK. DNA molecules are circularized at low concentrations that favor intramolecular ligation. Unwanted linear DNA is degraded with an exonuclease cocktail (comprised of Exonuclease I, Lambda exonuclease and Plasmid-Safe ATP-dependent DNase). Purified circular DNA is treated with Cas9:sgRNA RNP and cleaved DNA ends at on- and off-target sites are released for NGS library preparation, PCR amplification, and pair-end high-throughput sequencing.
