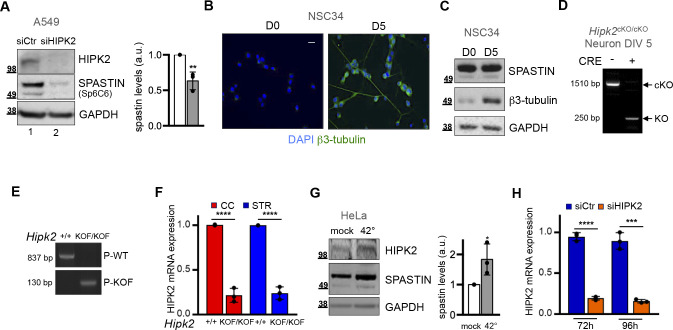
Figure S1. Spastin expression is regulated by HIPK2.
(A) Total cell extracts of A549 cells shown in Fig 1A were run twice and Western blot (WB) analysis performed by using the sp6C6 monoclonal spastin Ab for spastin detection. Representative WB and its quantification are reported. Statistical difference, unpaired t test. (B, C) NSC34 cells were incubated in differentiation medium. Differentiation was evaluated 5 d after treatment by IF and WB using β3-tubulin neuronal marker. In (B), representative images of β3-tubulin staining at indicated days after differentiation are reported. Scale bar, 10 μM. In (C), representative WB with indicated Abs is shown. (D) Indicated cortical neurons were analysed by PCR using primers amplifying cKO and KO Hipk2 alleles. The effect of CRE recombinase in Hipk2cKO/cKO is showed by the amplification of the KO allele. (E) PCR genotyping analysis of the genomic DNA extracted from tails derived from indicated mice by using primers specifically amplifying WT (P-WT) or KOF Hipk2 alleles (P-KOF). (F) HIPK2 mRNA residual levels were analysed by real time RT-PCR in tissues analysed in Fig 1F and G. Relative fold-change of HIPK2 mRNA levels, using actin mRNA as normalizer, are represented as mean ± SD of three independents. Statistical differences, relative to +/+ counterparts, unpaired t test. (G) HeLa cells were incubated in standard culture condition at 37°C (mock) or shifted to 42°C for 1 h to induce HIPK2 stabilization and analysed by WB with indicated Abs. Statistical difference, unpaired t test. (H) HeLa cells transfected as in Fig 1A were analysed by real-time RT-PCR at the indicated time post transfection. Relative fold-change of HIPK2 mRNA levels, using GAPDH mRNA as normalizer, are represented as mean ± SD of three independent experiments. Statistical differences, unpaired t test.