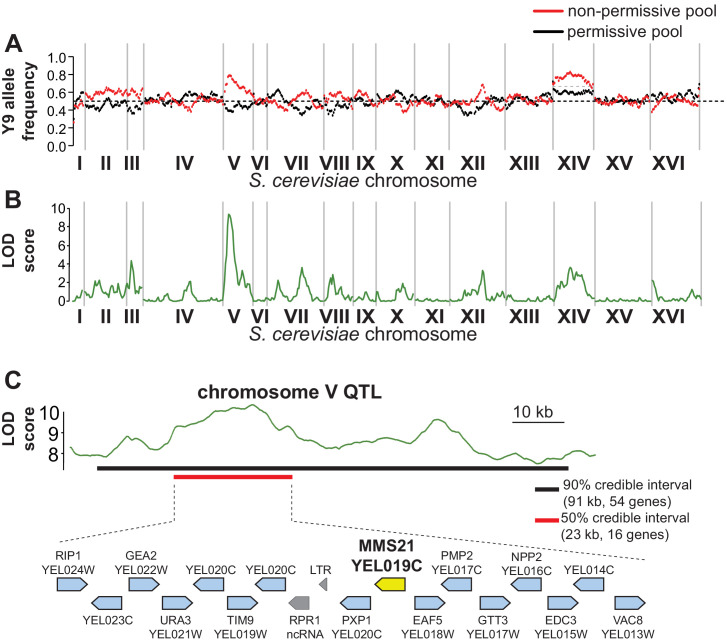
Figure 4. QTL mapping identifies a plasmid instability locus on Y9 chromosome V.
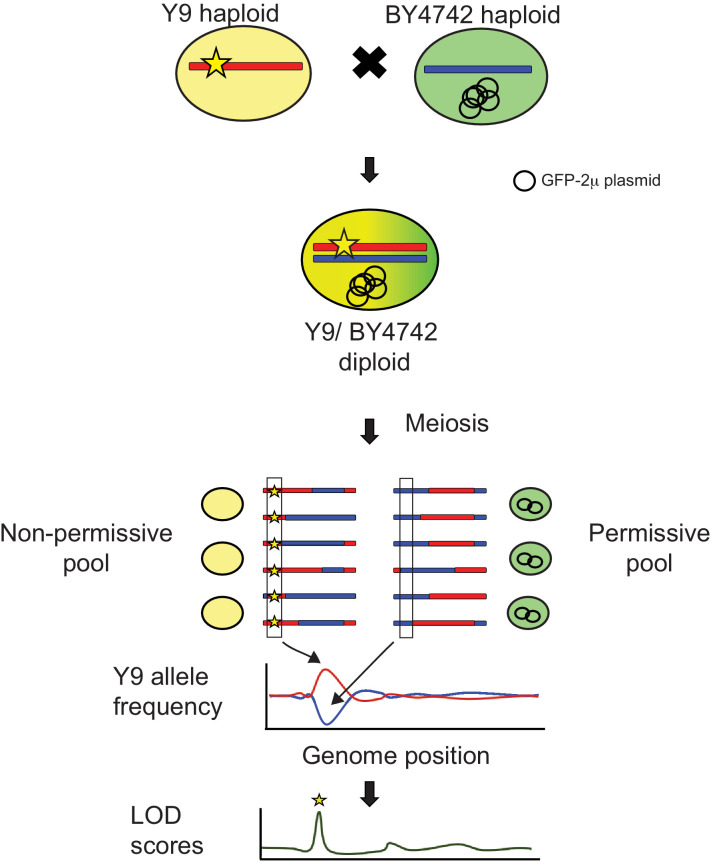
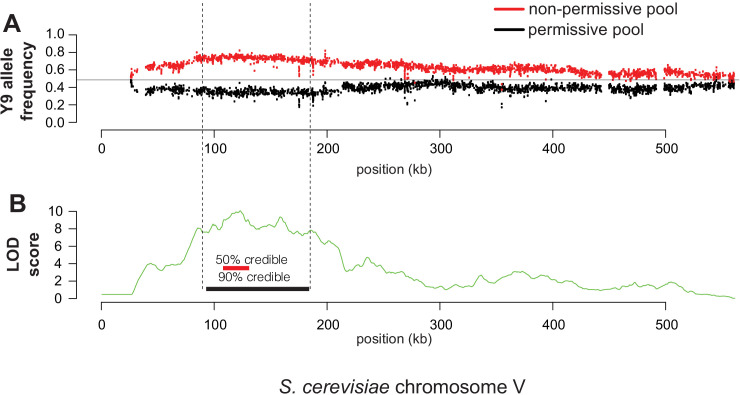
(A) We plotted the mean Y9 SNP allele frequency in 20 kb windows for the ‘non-permissive’ (red) and ‘permissive’ (black) pools of meiotic haploid progeny from BY4742/Y9 heterozygous diploid parents. Associations with a plasmid instability locus would show an increased representation of Y9 alleles in the non-permissive pool and a decreased representation of the BY4742 haplotype in the permissive pool (dotted line indicates equal representation). The increased representation of Y9 alleles on chromosome XIV in both pools is a result of a segregating disomy in the Y9 parent that we show does not affect the plasmid instability phenotype (Figure 4—figure supplement 2). (B) Based on the allele frequencies of individual SNPs, we used MULTIPOOL to calculate LOD scores for association with the plasmid instability phenotype. We observe a highly significant LOD score (10.00) on chromosome V. The peak is fairly sharp and reaches maximal LOD score at chrV:122.3–122.7 kb (sacCer3 coordinates). All loci have allele frequencies skewed in the expected direction; the restrictive pool is enriched for Y9 alleles. (C) MULTIPOOL 90% (54 genes, 91.2 kb, chrV:92.4–183.6 kb) and 50% (16 genes, 23.1 kb, chrV:107.3–130.4 kb) credible intervals for the chromosome V QTL. Among the 16 genes in the 50% credible interval is MMS21, which encodes an essential SUMO E3-ligase.