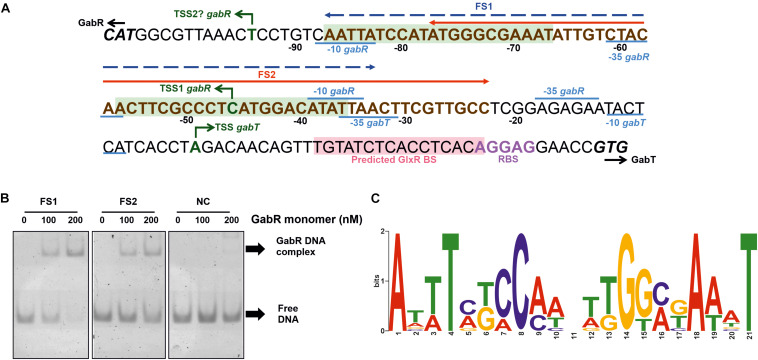
FIGURE 6.
Determination of the binding site(s) of GabR in the gabTDP promoter region. (A) Sequence of the gabR-gabT intergenic region. The ATG start codon of gabR (cg0565) and the GTG start codon of gabT (cg0566) are shown in bold and italic. The transcriptional start sites of gabR and gabT are indicated with green arrows. The corresponding –10 and –35 regions are labeled in blue. The ribosome binding site of gabTDP is labeled in purple, and the predicted GlxR binding site is marked with light red background. The binding region for GabR determined in this work by EMSAs is marked in bold brown letters (compare Supplementary Figure 3). Fragments FS1 (–87 to –33) and FS2 (–77 to –23) are marked in the figure by a blue or red arrow, respectively. Potential binding sites for GabR are marked with light green background. (B) EMSAs with purified GabR and DNA fragments FS1 and FS2. The fragments were incubated with the indicated GabR concentrations (given in nM of monomers). A 55 bp fragment downstream of the predicted binding sites was used as negative control (NC). (C) Proposed GabR consensus binding site identified with MEME software using the gabR-gabT intergenic regions of C. glutamicum WT, Corynebacterium deserti and Corynebacterium callunae as input sequences.