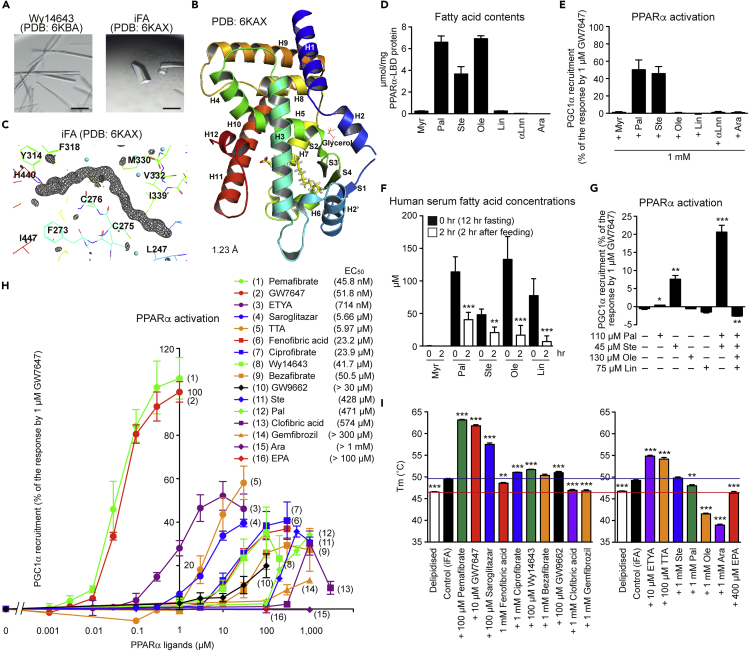
Figure 1.
Structures of Intrinsic Fatty Acid-Bound PPARα-LBD and PPARα Activation by Endogenous Fatty Acids
(A) Intrinsic fatty acid (iFA)-bound crystals (right) obtained using crushed Wy14643-bound crystals (left) as crystal nuclei. Scale bars: 100 μm.
(B) High-resolution (1.23 Å) structure of PPARα-LBD and iFA (yellow). α-helices, β-sheets, and glycerol are labeled.
(C) Magnified view of iFA and surrounding amino acids. The electron density is shown in the mesh by Fo-Fc omit map contoured at 3.0σ. Water molecules are presented as cyan spheres.
(D) Contents of the seven major fatty acids (of E. coli origin) in PPARα-LBD proteins measured by LC-MS/MS analysis.
(E) PPARα activation (PGC1α coactivator recruitment) by endogenous fatty acids. Maximal response by 1 μM GW7647 was set as 100 (%).
(F) Serum fatty acid contents of five healthy male volunteers who were fasted for 12 h and then fed. Serum fatty acid levels were decreased after 2 h.
(G) Effects of fatty acid combinations on PPARα activation.
(H) Concentration-dependent PPARα activation by several ligands; their EC50 values are shown in parentheses.
(I) Effects of several ligands on PPARα-LBD thermostability. Ligands were dissolved in 100% DMSO (left) or 100% ethanol (right), and their final concentrations in assays were 0.1%.
The differences versus control (0 h in (F), no lipids in (G), and iFA-bound proteins in (I)) are significant at ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001. Data are means ± SEM in (D) (n = 3), (E) (n = 3), (G) (n = 4), (H) (n = 3–4), and (I) (n = 4–6), and means ± SD in (F) (n = 5). Myr, myristic acid; Pal, palmitic acid; Ste, stearic acid; Ole, oleic acid; Lin, linoleic acid; αLnn, α-linolenic acid; and Ara, arachidonic acid.