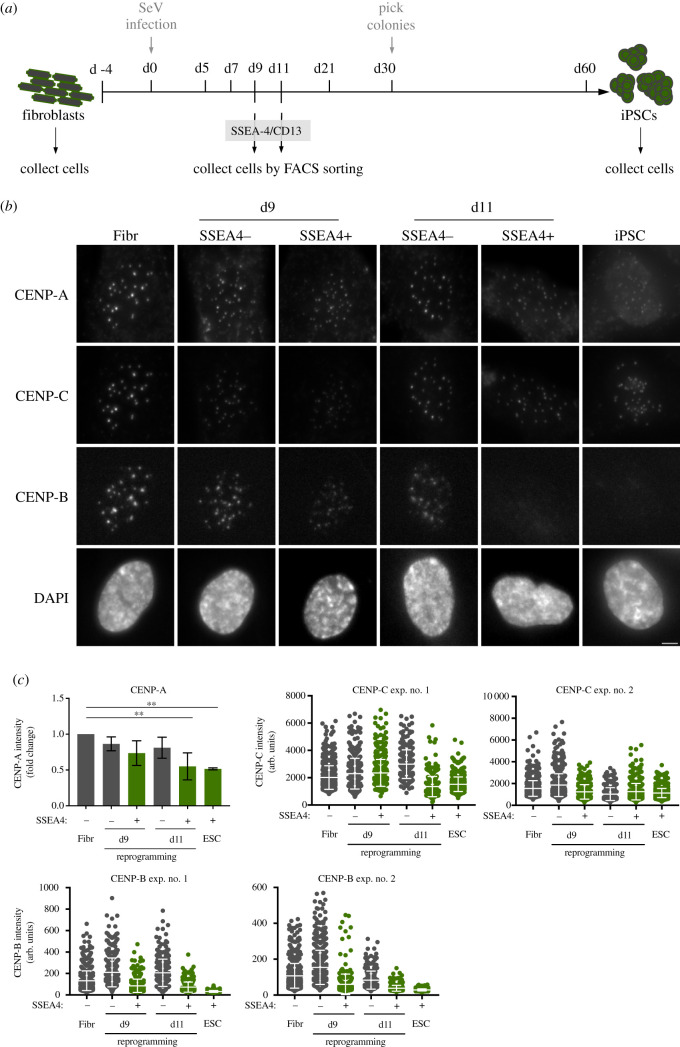
Figure 5.
CENP-A loss is induced during early reprogramming of fibroblasts to iPSCs. (a) Outline of the general strategy to reprogramme iPSCs from fibroblasts: human primary fibroblasts are reprogrammed by infection with Sendai virus (SeV) expressing Oct4, Sox2, Klf4 and c-Myc. At days 9 and 11 after infection (d9 and d11, respectively), cells are incubated with antibodies specific for SSEA-4 (early pluripotency marker) and CD13 (fibroblast marker) and collected by FACS sorting. Thirty days after infection, visible colonies appear and can be picked under the microscope. Single colonies are picked, expanded and kept in culture. The cells collected at days 9 and 11, the initial fibroblast population and fully reprogrammed iPSCs (reprogrammed from those fibroblasts), were stained for CENP-A, CENP-B and CENP-C and counterstained with DAPI. (b) Representative immunofluorescence images from cells collected by FACS at d9 and d11 and sorted by pluripotency profile (SSEA4 negative and CD13 positive–refractory to reprogramming–versus SSEA4 positive and CD13 negative cells–prone to reprogram) and control cells. (c) Quantification of centromere intensities as shown in (b). Average centromere intensities were determined using automatic centromere recognition and quantification (CRaQ; see methods) for indicated cell types. For CENP-A, the average and standard error of the mean of three replicate experiments is shown for indicated cell types. Centromere intensities are normalized to those of fibroblasts. For CENP-B and CENP-C, two experiments were performed and analysed. Horizontal lines represent the mean, and whiskers represent standard deviation, for each sample. Scale bar = 2 µm.