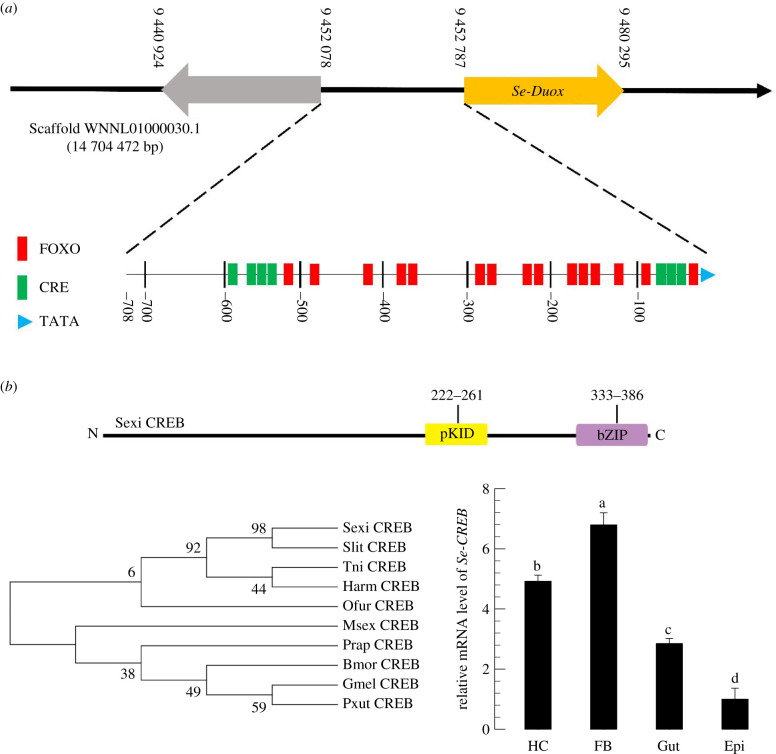
Figure 5.
Identification of CRE and CREB associated with Se-Duox. (a) Promoter analysis of dual oxidase gene of S. exigua using PROMO and GPMiner programs. The upstream region from ATG start codon of Se-Duox was analysed for promoter components. (b) Functional domain analysis and phylogenetic analysis of Se-CREB with other lepidopteran CREBs based on their amino acid sequences. Domains were predicted using HMMER (https://www.ebi.ac.uk) and Pfam (http://pfam.xfam.org). Predicted domains include ‘pKID’ for phosphorylated kinase-inducible-domain and ‘bZIP’ for basic leucine zipper. The tree was generated by the neighbour-joining method using MEGA 6.0. Bootstrapping values were obtained with 1000 repetitions to support branch and clustering. Amino acid sequences were retrieved from GenBank. Accession numbers of genes are shown in electronic supplementary material, table S3. (c) Expression patterns in indicated tissues of L5D2 larvae, including haemocyte (HC), fat body (FB), midgut (Gut) and epidermis (Epi). A ribosomal gene, RL32, was used as a reference gene. Each treatment was replicated three times with independent tissue preparations. Different letters above standard deviation bars indicate significant differences among means at Type I error = 0.05 (LSD test).