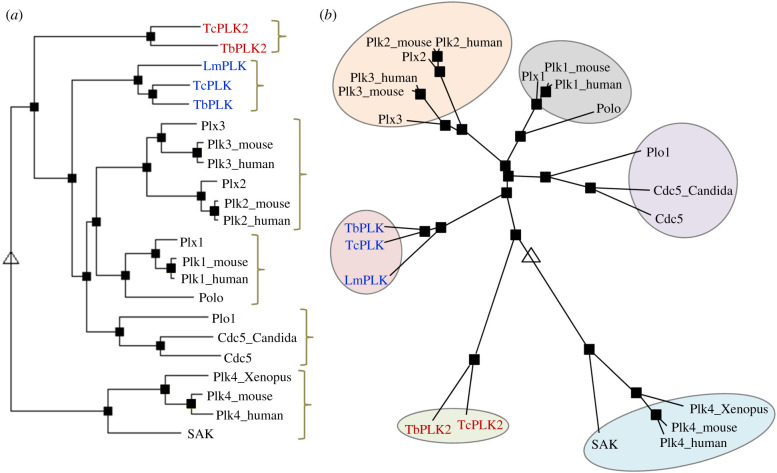
Figure 2.
Phylogenetic analysis of Plk family members in diverse organisms. (a) Full-length amino acid sequences of known Plk family members from different organisms were aligned using hierarchical clustering, and a rooted tree was constructed based on the sequence alignment obtained using the MultAlin server (http://multalin.toulouse.inra.fr/multalin/). Plks from the following organisms were used: H. sapiens (Plk1_human−Plk4_human; accession nos: P53350, Q9NYY3, Q9H4B4 and O00444); Mus musculus (Plk1_mouse−Plk4_mouse; accession nos: Q07832, P53351, Q60806 and Q64702); Xenopus laevis (Plx1−Plx3 and Plk4_Xenopus; accession nos: P70032, Q90XS4, Q90XS3 and Q6PAD2); D. melanogaster (Polo and SAK; accession nos: P52304 and O97143); Sacchromyces cerevisiae (Cdc5; accession no.: P32562); Schizosacchromyces pombe (Plo1; accession no.: P50528); Candida albicans (Cdc5_Candida; accession no.: Q5ABG0); T. brucei (TbPLK and TbPLK2; accession nos: Tb927.7.6310 and Tb927.6.5100); Trypanosoma cruzi (TcPLK and TcPLK2; accession nos: TcCLB.506513.160 and TcCLB.506945.350); L. major (LmPLK; accession no.: LmjF.17.0790). (b) A rooted radial tree shows the distance relationship among the Plks from different organisms. The tree was generated using the same hierarchical clustering as in (a). The triangle indicates the root of the tree, and the squares indicate branch points.