Figure 2.
ERK Signaling Is Required for Maximal Nr4a Expression in T Cells, but Nr4a3 Can Be Activated by NFAT Overexpression Alone
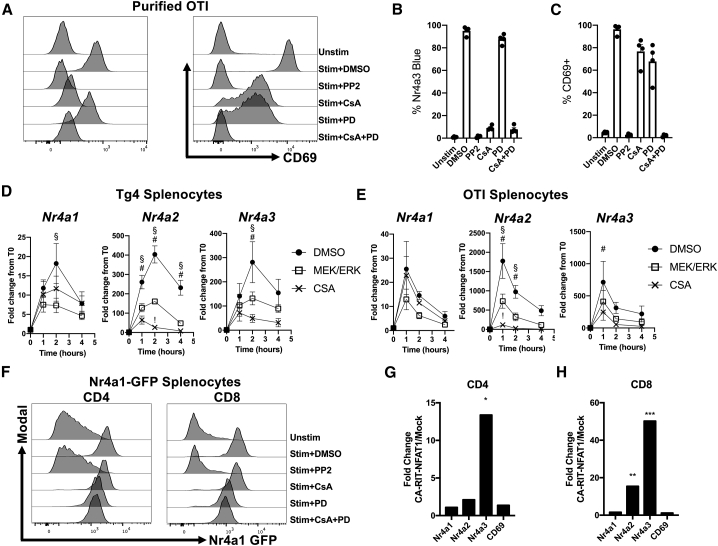
(A–C) Purified OTI Nr4a3-Tocky CD8+ T cells were stimulated with 1 μM ova peptide for 4 h in the presence of DMSO, 10 μM PP2, 1 μM CsA, 5 μM PD0325901 (PD), or CsA and PD. Cells were analyzed by flow cytometry for the expression of Nr4a3-Timer Blue and CD69 in live CD8+ T cells. Summary data from %Nr4a3-Timer Blue+ (B) and %CD69+ (C); n = 4. Bars represent mean ± SEM, dots represent individual donor mice. (D–E) Splenocytes from Tg4 Nr4a3-Tocky mice were stimulated with 10 μM 4Y-MBP (D) or OTI Nr4a3-Tocky mice with 1 μM ova peptide (E) for 4 h in the presence of DMSO, 1 μM CsA, or 5 μM PD; RNA was extracted at 0, 1, 2, and 4 h following culture; and fold change in Nr4a1–Nr4a3 transcription was measured. DMSO treated, black circles; CsA treated, black crosses; or PD treated, open squares; n = 4 (D); or n = 3 (E). Bars represent mean ± SEM; #, CsA treated significantly reduced (p < 0.05) from DMSO; §, PD treated significantly reduced (p < 0.05) from DMSO; !, CsA treated significantly reduced (p < 0.05) from PD. Statistical analysis by two-way ANOVA with Tukey’s multiple comparisons test.
(F) Splenocytes from Nr4a1-GFP mice were cultured for 4 h with 5 μg/ml anti-CD3 or media alone (unstim) in the presence of 0.1% DMSO, 10 μM PP2, 1 μM CsA or 5 μM PD, or 1 μM CsA and 5 μM PD. Live CD4+ (left) or CD8+ (right) T cells were then analyzed for Nr4a1-GFP expression.
(G and H) Analysis of RNA-seq data from GEO: GSE64409 displaying the fold change in expression of Nr4a1, Nr4a2, Nr4a3, and Cd69 in CD4+ (G) or CD8+ (H) T cells transduced with CA-RIT-NFAT1. Adjusted p values based on DESeq analysis; ∗p = 0.0206, ∗∗p = 0.00305, ∗∗∗p = 0.00016. See also Figure S2.