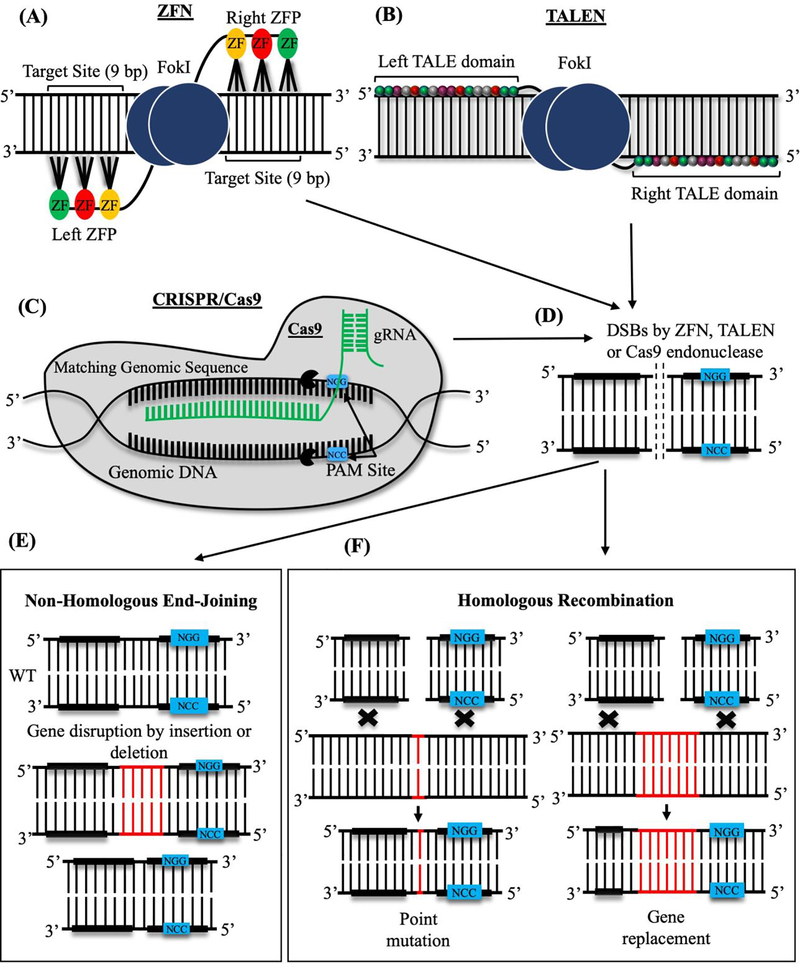
Figure 1: Site-specific genome editing methods that use endonuclease activity.
(A) The Zinc-Finger Nuclease complex (ZFN) consists of the FokI endonuclease domain (blue) and several Zinc finger (ZF) DNA-binding domains (yellow, red and green). Each ZF domain recognizes a 3-base pair (bp) DNA sequence. (B) Transcription Activator-Like Effector Nuclease (TALEN) is shown as a fusion protein consisting of FokI endonuclease domain and TALE DNA-binding domains. Sixteen colored subunits in each TALE domain recognize a single base pair of DNA sequences. (C) CRISPR/Cas9 system is comprised of Clustered Regulatory Interspaced Short Palindromic Repeats (CRISPR) and CRISPR associated (Cas) protein 9. Guide RNA (gRNA, green) directs Cas9 endonuclease (grey) to specific DNA sites, which are located close to short Protospacer Adjacent Motifs (PAM, blue). Cas9 cleaves DNA upstream of PAM. (D) FokI and Cas9 endonucleases cause double-strand DNA breaks (DSBs). DSBs are repaired through either NHEJ or HR mechanisms. (E) NHEJ repair is based on nucleotide deletions or insertions into cleaved DNA. (F) Repair through HR mechanism occurs in the presence of the donor DNA template, which contains homology regions on both sides of the template. Exogenous DNA is specifically inserted into the target site to modify the gene.