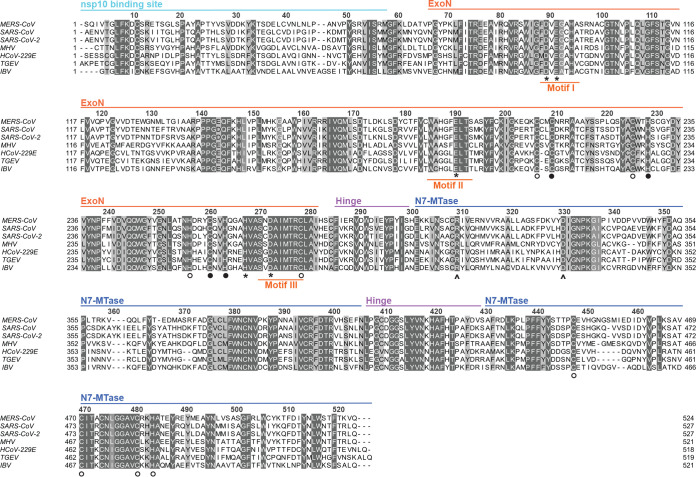
FIG 1.
Alignment of nsp14 amino acid sequences from selected coronaviruses. Sequences of the ExoN and N7-MTase domains in MERS-CoV (NC-019843), SARS-CoV (NC_004718), SARS-CoV-2 (NC_045512.2), MHV (NP_045298), HCoV-229E (NC_002645), TGEV (AJ271965), and IBV (NP_040829) were used for the analysis. The domains indicated at the top are based on the SARS-CoV-nsp14 secondary structure (PDB 5NFY) (21). Fully conserved residues are shown with white letters on dark gray (above 70% conservation), whereas partially conserved residues are displayed with lighter shades of gray. Catalytic residues and residues involved in formation of zinc fingers are marked with asterisks and circles, respectively. Filled circles indicate zinc fingers targeted by mutagenesis (Fig. 2A), while arrowheads identify the two N7-MTase domain residues mutated to generate the MTase negative control used in biochemical assays. The alignment was generated using Clustal Omega (104) and edited using Jalview version 2.11 (105).