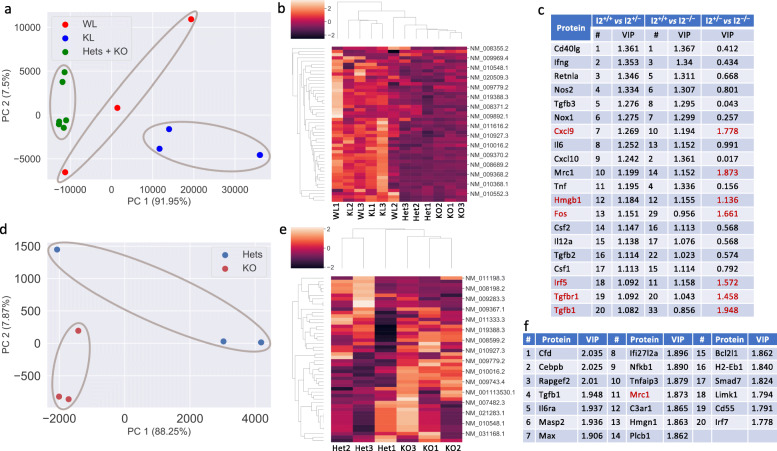
Fig. 6.
Bioinformatics analysis of NanoString data for livers of Aldh1l2+/+ and Aldh1l2−/− male mice. PCA was carried out using the sklearn package in Python. PLS-DA analysis was carried out using the package mixOmics in R. a PCA for wild type (WL), Aldh1l1−/− (KL), Aldh1l2+/− (Hets), and Aldh1l2−/− (KO) mice. Raw data were first normalized and then PCA was applied. No scaling was used prior to PCA. PC1 and PC2 explain 91.95% and 7.5% of the total variance of the data, respectively. b Heat map representation of the NanoString data for the four genotypes (see Supplementary Fig. S11 for the full-size image). Each protein was standardized so that it has mean 0 and standard deviation 1. c Top 20 discriminating proteins for Aldh1l+/− and Aldh1l2−/− from wild type mice according to PLS-DA VIP values. Most of these genes do not discriminate between Aldh1l2+/− and Aldh1l2−/− genotypes. d PCA for Aldh1l2+/− (Hets) and Aldh1l2−/− (KO) mice. Raw data were first normalized and then PCA was applied. No scaling was used prior to PCA. PC1 and PC2 explain 88.25% and 7.87% of the total variance of the data, respectively. e Heat map representation of the NanoString data for the Aldh1l+/− and Aldh1l2−/− genotypes (based on the entire panel of 242 genes; see Supplementary Fig. S12 for the full-size image). Each protein was standardized so that it has mean 0 and standard deviation 1. f Top 20 discriminating proteins for Aldh1l+/− versus Aldh1l2−/− genotype according to PLS-DA VIP values