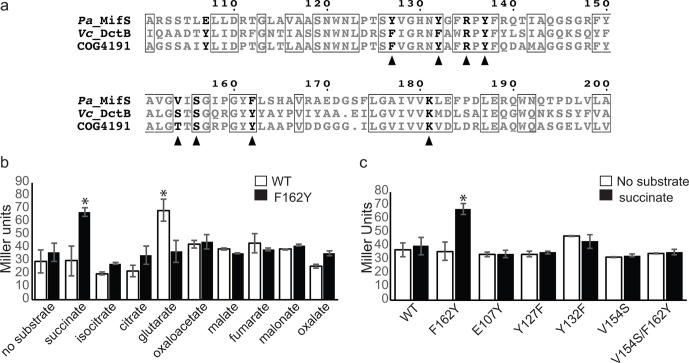
Fig. 5.
MifS sensor domain residue Phe162 required for substrate specificity. (a) Alignment of substrate binding pocket sequences of MifS, Vc_DctB and COG4191 (DctB family consensus sequence). Residues conserved in all three sequences are shown in boxes. Residues required for α-KG response in MifS are shown in bold. Residues required for succinate binding in Vc_DctB are indicated with a triangle. Alignment was generated using clustal Omega [56] and displayed using ESPript 3.0 [57]. (b) E. coli cells harbouring the wild-type mifS gene or the mifSF162Y gene along with the mifR gene and PPA5530::lacZ reporter were grown in LB media to an OD600 value of 0.3 and induced with a panel of dicarboxylates at a final concentration of 20 mM. (c) Wild-type MifS or a MifS variant with a mutation resembling the Vc_DctB binding pocket was expressed in E. coli cells harbouring the mifR gene and the PPA5530::lacZ reporter. Cells were grown in LB media to an OD600 value of 0.3 and challenged with succinate or no substrate. LacZ expression levels were measured 60 min post-induction. Data points represent mean values±the standard deviations (n=3). Analysis of variance was performed by using Dunnett’s post-hoc test (α value of 0.05) to identify significant differences (P<0.0001; marked with an asterisk).