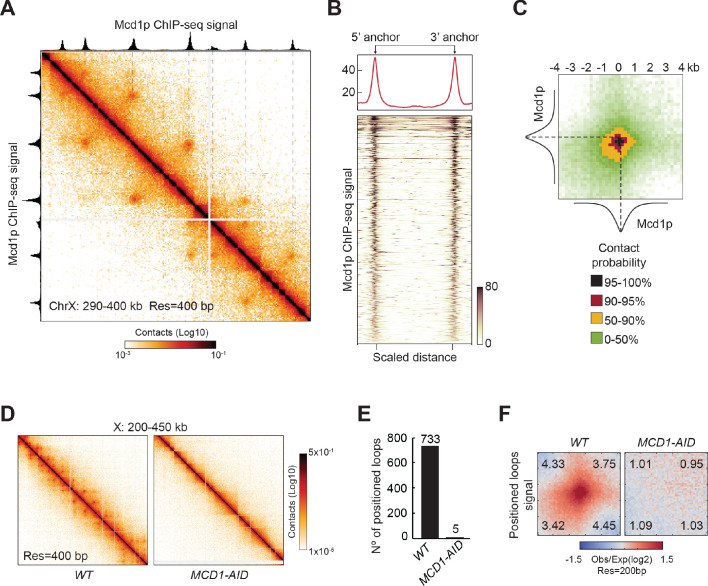
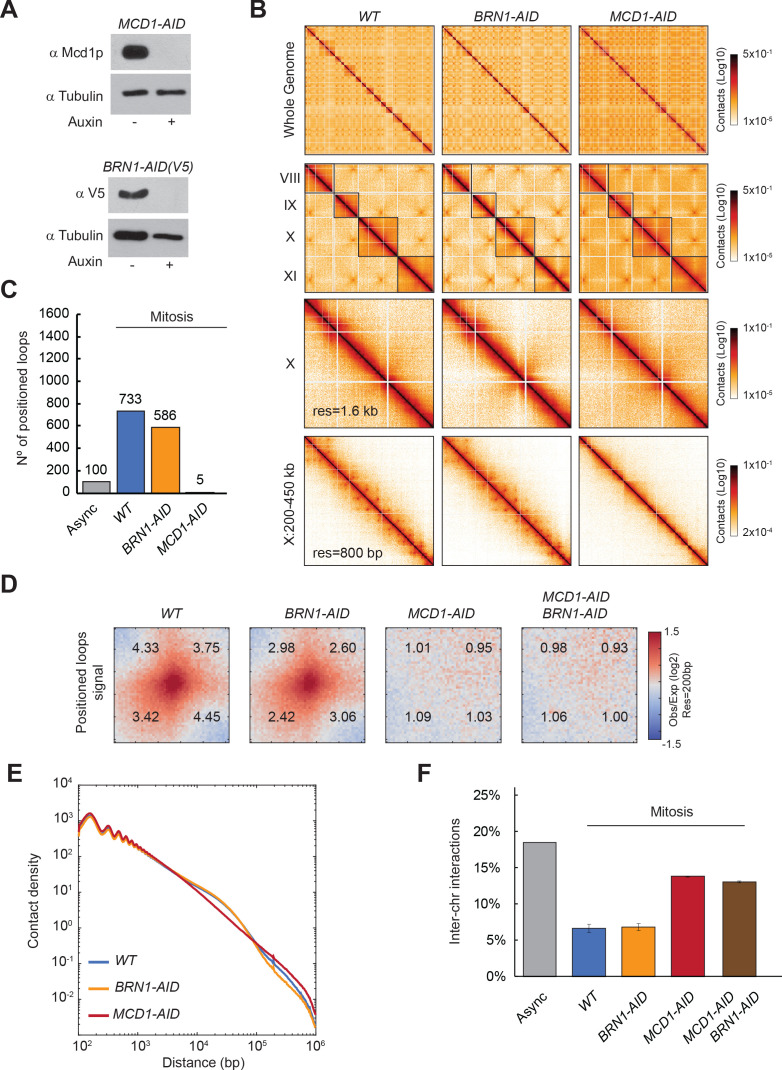
Figure 2. Mitotic positioned loops are cohesin-dependent.
(A) Cohesin peaks colocalize with the loop anchors of positioned loops. Contact map showing the interactions in the 290–400 kb region of chromosome X overlays with Mcd1p ChIP-seq tracks on top and left. The dashed lines indicate the colocalization of anchors of positioned loops and Mcd1p peaks. (B) Cohesin binding is enriched at anchors for positioned loops genome-wide. Heatmap (bottom panel) shows the enrichment of the Mcd1p ChIP-seq reads at the loop anchors genome-wide (732 rows). Pairs of loop anchors were rescaled to the same length. Genome-wide average ChIP-seq signal over the rescaled loop anchors is plotted (top panel). (C) Mcd1p peaks center on the anchors for positioned loops. Heatmaps were plotted with 200 bp resolution of contact map signal in ±4 kb regions surrounding the paired Mcd1p peaks. The contact map is colored by the contact probability with >95% in black, 90–95% in red, 50–90% in yellow, 0–50% in shades of green. The average Mcd1p peak is overlaid with contact maps and centered at the loop bases. (D) The positioned loop signal on the contact map is lost upon cohesin depletion. Contact maps for WT and MCD1-AID Micro-C data show substantial loss of loop signal over chromosome X: 200–450 kb upon Mcd1p degradation. (E) Positioned loops are gone upon Mcd1p-depletion. The bar chart shows the loop number called by the HiCCUPS program in WT (733) and MCD1-AID (5). (F) Genome-wide analysis confirms that the positioned loop signal is gone in MCD1-AID cells. Heatmaps were plotted with 200 bp resolution data for WT and MCD1-AID in ±5 kb regions surrounding the wild-type loop anchors.