Figure 3.
Heterozygous PRS Enhancer Deletion In Vitro Affects SOX9 Expression during a Restricted Window of Development
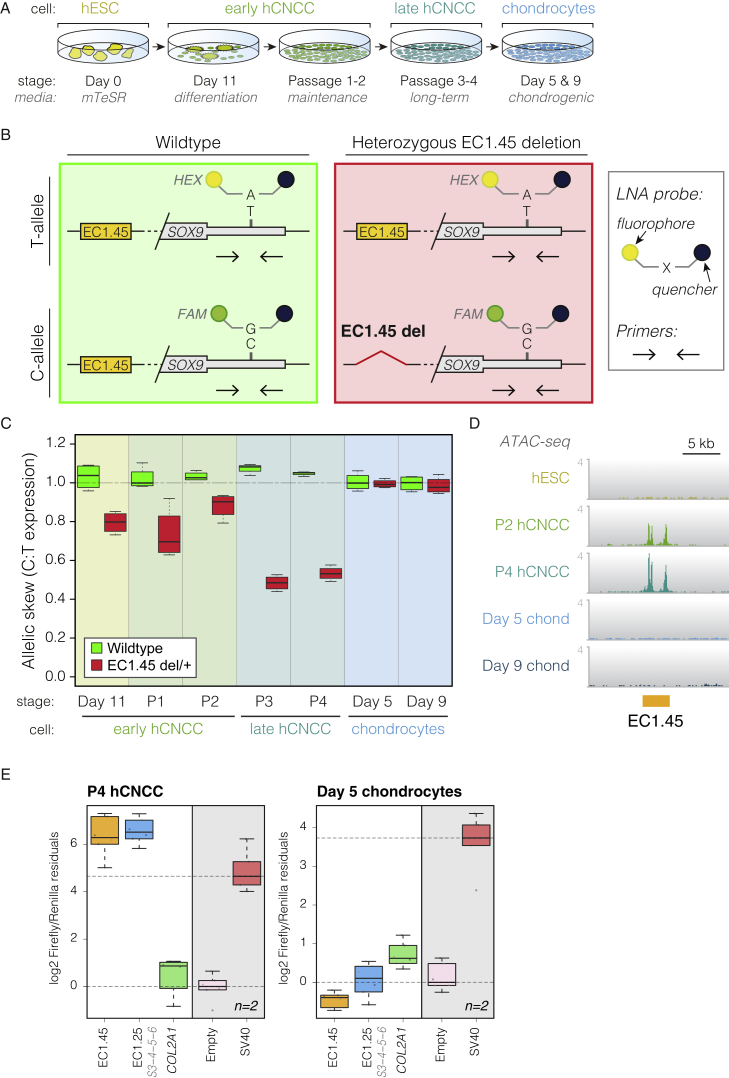
(A) Overview of differentiation, including early hCNCCs at day 11, passage 1–2 early hCNCCs, passage 3–4 late hCNCCs, and chondrocytes on days 5 and 9.
(B) Schematic of allele-specific RT-ddPCR, indicating primers and LNA probes (HEX/FAM) for the T/C SNP (rs74999341) in the SOX9 3′ UTR. Shown are wild-type (left) and heterozygous EC1.45 deletion (right).
(C) RT-ddPCR for wild-type (green boxplot) and EC1.45 heterozygous deletion (red), plotting SOX9 C:T expression ratio.
(D) ATAC-seq reveals hCNCC-specific accessibility for EC1.45. Shown are representative traces from 3–4 replicates.
(E) Luciferase assay for late hCNCCs (left) and chondrocytes (right). A COL2A1 enhancer is active in both cell types, whereas EC1.45 and EC1.25 become inactive in chondrocytes.