Figure 4.
Dissection of EC1.45 Enhancer Region Uncovers a Core Role of the Coordinator Motif and TWIST1 Binding in Developmental Enhancer Regulation
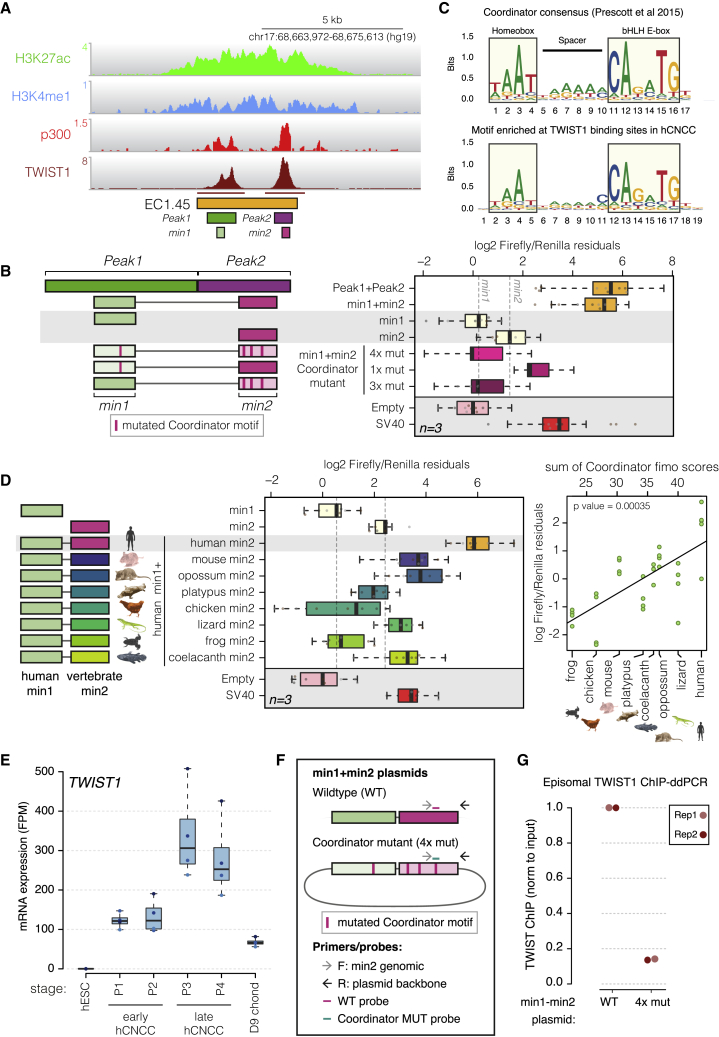
(A) TWIST1 ChIP-seq peaks (marked under track) at EC1.45 overlap p300 Peak1 and Peak2 and minimally active sequences (min1 and min2).
(B) Luciferase assay for EC1.45 min1 and min2, tested separately and combined, along with Coordinator mutant sequences. Left: schematic of the constructs.
(C) Coordinator motif (top; Prescott et al., 2015) compared with the motif enriched at TWIST1 binding sites in hCNCCs (bottom).
(D) Luciferase assay for the heterologous enhancer sequence for human min1 plus vertebrate min2. Left: schematic of the constructs. A scatterplot depicts the luciferase signal compared with the sum of Coordinator scores (ANOVA p = 0.00035; right).
(E) TWIST1 is upregulated during hCNCC differentiation and reduced in chondrocytes (fragments per million [FPM]).
(F) Schematic of plasmids, primers, and probes for ChIP-ddPCR for wild-type (WT) and Coordinator mutant (4x mut) min1+min2 plasmids. F, forward; R, reverse.
(G) TWIST1 ChIP-ddPCR for P4 late hCNCCs transfected with the plasmids in (F), normalized to input, and WT adjusted to 1. Two biological replicates are depicted.
See also Figure S5.