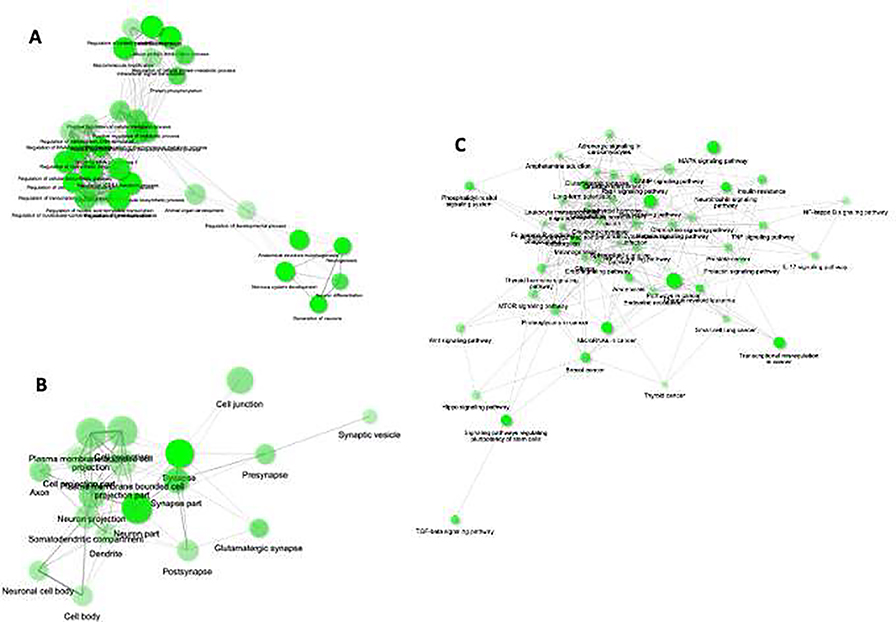
Fig. 2: Gene ontology and pathway prediction.
GO terms for A) biological process and B) cellular component based on predicted 1,092 target genes. The GO prediction analysis was done following an FDR corrected p-value cutoff 0.05 to determine the gene set enrichment in biological process and cellular component category separately by ShinyGO program. Each category the enriched terms were used to create networks where nodes are presented with terms and connected with edges. As shown in the graphs, if two nodes are connected, then they share 20% (default) or more genes. Bigger nodes represent larger gene sets. Thicker edges represent more overlapped genes. C) A pathway-based network map (p value cutoff <0.05 [FDR], Edge Cutoff: 0.2; most significant display terms: 50) was prepared showing the predicted involvement of various neuro-molecular pathways, those are earlier reported to be important in MDD pathogenesis.