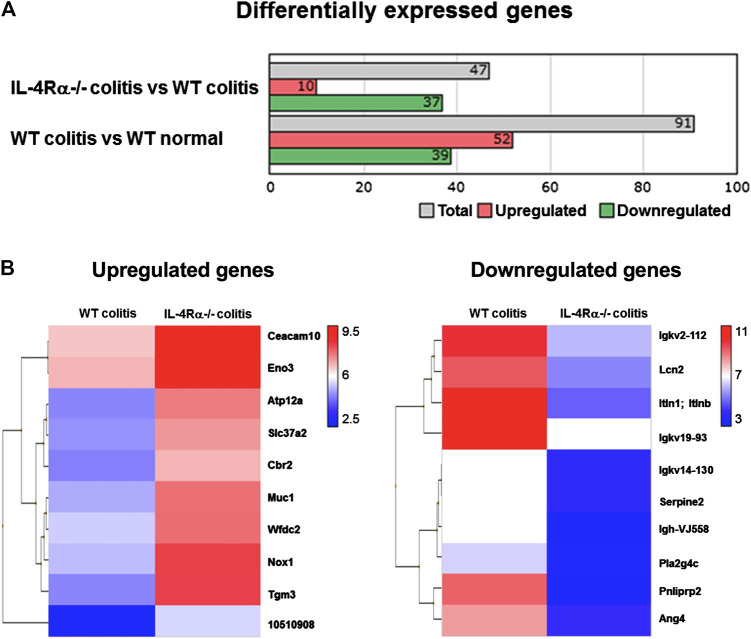
FIGURE 4.
Transcriptomic analysis shows that IL-4Rα-/- mice exhibit a high level of Nox1 mRNA expression in DSS-induced colitis model. Microarray data sets of WT normal, WT colitis (day 7) and IL-4Rα-/- colitis (day 7) mice are analyzed using Transcriptomic Analysis Console (TAC) Software. (A) Differentially expressed genes whose expression levels are more than four-fold or less than quarter in comparison between IL-4Rα-/- colitis and WT colitis or WT colitis and WT normal. (B) Heat map shows top 10 upregulated (left) and downregulated (right) in IL-4Rα-/- colitis mice compared with WT colitis mice. The color range represents log2-transformed fold changes. Each row corresponds to each gene, and each column corresponds to each group. Genes were sorted using hierarchical clustering based on Euclidean distance and average linkage, as shown in the dendrogram on the left side of the heatmap. The 10 gene names are displayed on the right side of the heatmap.