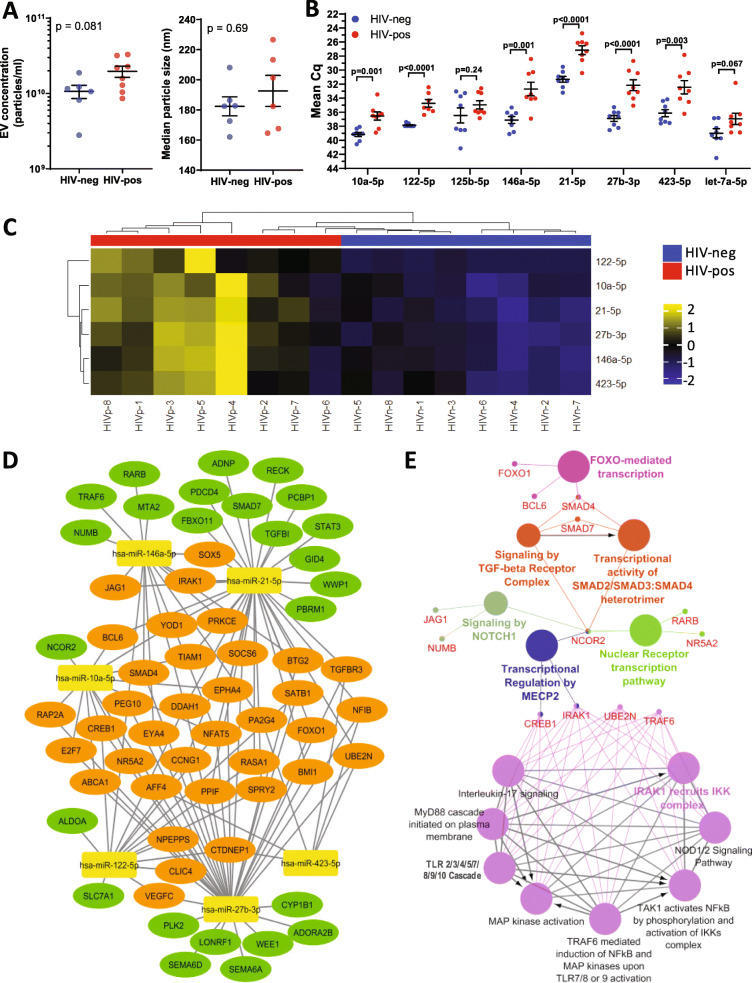
Fig. 6.
qPCR validation of candidate miRNAs and functional associations of differentially expressed miRNAs. a EV concentration (left) and median size (right) measured by NTA in exosome fractions isolated from HIV-positive and HIV-negative subjects from the validation cohort. b Comparison of Cq values of candidate miRNAs isolated from plasma EVs of HIV-positive and HIV-negative subjects using miRCURY LNA RT-qPCR kits. Mean and SEM are shown. Significance was calculated using Mann Whitney test. (n = 8 HIV-positive and n = 8 HIV-negative subjects). c Heatmap shows unsupervised hierarchical clustering of miRNAs (n = 6) that distinguish HIV-positive from HIV-negative subjects (p < 0.05, Mann-Whitney test). d . miRNA target–gene network was constructed using Cytoscape for 61 target genes of 6 differentially expressed miRNAs in the validation cohort. miRNAs are depicted by yellow nodes. 36 genes (60%) are targeted by more than one miRNA (orange nodes); genes targeted by one miRNA are depicted by green nodes. e Functionally grouped network of enriched categories was generated for miRNA-target genes with REACTOME pathways using the ClueGO plugin in Cytoscape as described in the Methods. Node sizes are indicative of p-values (larger size corresponds to smaller p-values and vice versa). Nodes belonging to one functional group share the same color. Similar pathway terms with identical genes were fused and pathway term ‘TP53 regulation of cell cycle genes’ was omitted for clarity. Complete list of pathway terms and associated genes is shown in Supplementary Table 6