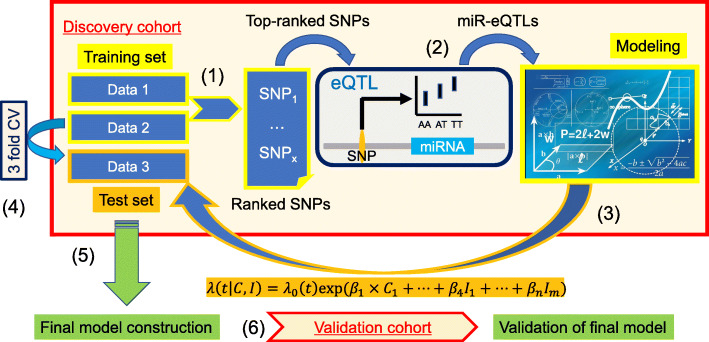
Fig. 1.
Construction workflow of our prognosis prediction model. We generated data sets of pre-selected SNPs based on the P values from logistic regression models. Two thirds of the entire discovery cohort was used to calculate the P values in each cross-validation step (1). Using the top-ranked SNPs, we focused on SNP-miRNA pairs with eQTL effects (miR-eQTLs) (2). Using a combination of miR-eQTLs and clinical factors, we constructed a prognosis prediction model based on a Cox proportional hazard model using two thirds of the discovery cohort. The adjusted model was then evaluated using the remaining one third (3). On the basis of the average C-index from the 3-fold cross-validation, we determined the optimal pre-selected SNPs for model construction (4). The final model was constructed using the entire discovery cohort (5), and the adjusted model was evaluated in the independent validation cohort (6)