Figure 2.
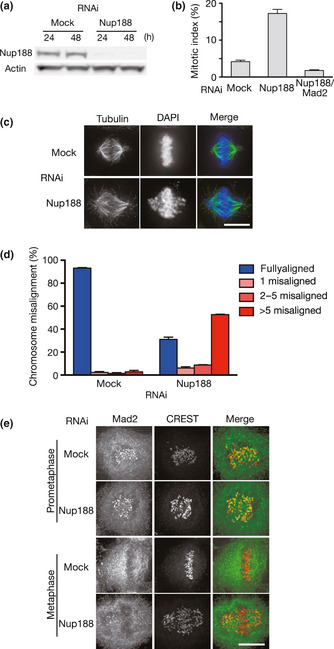
Nup188 depletion causes chromosome misalignment. (a) Depletion of Nup188 with a siRNA. Total cell lysates prepared from HeLa cells treated with mock or Nup188 siRNA for the indicated periods were separated by SDS‐PAGE and probed using western blotting with anti‐Nup188 or anti‐actin antibody. (b) Increased mitotic index in Nup188‐depleted cells. The mitotic index for mock‐, Nup188 siRNA‐ or Nup188/Mad2 siRNA‐treated HeLa cells was measured 48 h after transfection. Error bars represent the SD. (c) Nup188‐depleted cells showed chromosome misalignment. HeLa cells treated with mock or Nup188 siRNA for 48 h were stained with an anti‐tubulin antibody (green). DNA was stained with DAPI (blue). Bar, 10 μm. (d) Quantitative analysis of chromosome misalignment in Nup188‐depleted cells. HeLa cells were treated with mock or Nup188 siRNA for 48 h and with MG132 (10 μM) for the final 2 h. The number of misaligned chromosomes per cell was scored. Error bars represent the SD. (e) Mad2 localizes to kinetochores in Nup188‐depleted cells. HeLa cells treated with mock or Nup188 siRNA for 48 h were stained with an anti‐Mad2 antibody (green) and a CREST serum (red). Bar, 10 μm.
