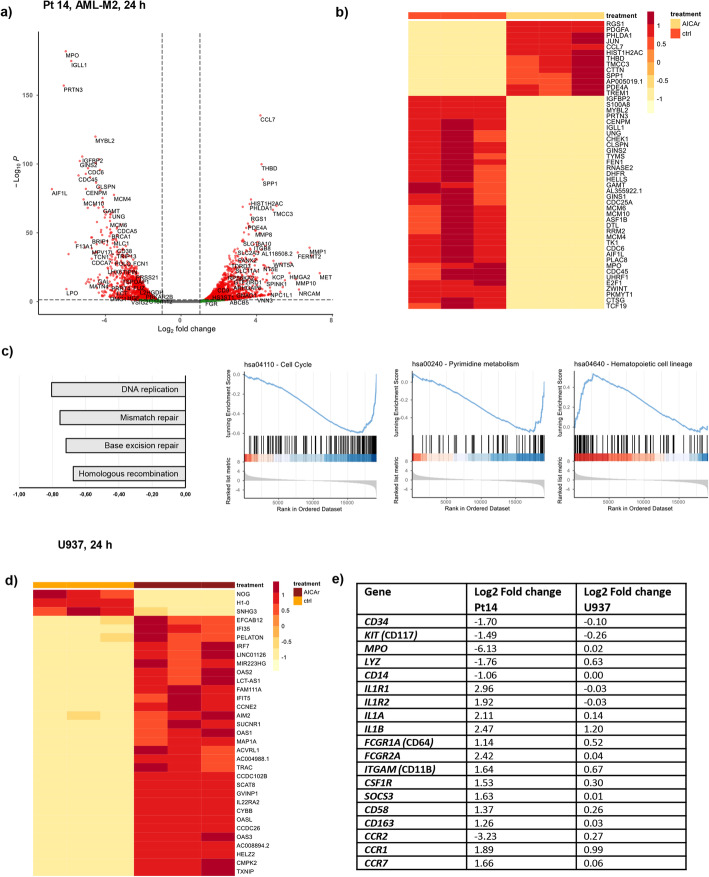
Fig. 4.
Differential gene expression in primary sample from AML-M2 patient (normal karyotype, FLT3wt NPM1wt, primary refractory) and U937 cells in response to AICAr. Bone marrow sample from the AML patient (Pt 14) in which AICAr induced differentiation in vitro and U937 cells were seeded in triplicates and treated with AICAr for 24 h. Differential gene expression was determined using RNA sequencing and DESeq2 analysis was performed. a) Volcano plot. The red dots: |log2 FC| ≥ 1, p.adjusted < 0.05; the green dots: |log2 FC| ≥ 1, p.adjusted > 0.05; the black dots: |log2 FC| ≤ 1, p.adjusted > 0.05. b) Heatmap from hierarchical clustering of top 50 significant genes ranked by p.adjusted value between untreated and AICAr-treated primary samples. c) Gene Set Enrichment Analysis (GSEA) for KEGG pathways: Top 4 most enriched pathways and GSEA plots (score curves) for pathways Cell Cycle, Pyrimidine Metabolism and Hematopoietic Cell Lineage. p.adjusted < 0.01. d) Heatmap from hierarchical clustering of top 30 significant genes ranked by p.adjusted value between control and AICAr-treated U937 cells. e) Log2Fold changes in the expression of genes associated with immature cells, monocytes and macrophages in primary sample and U937 cells