Figure 6.
Cohesin Negatively Regulates Memory for Most Genes in the GBP and HLA Clusters but Not for Genes Outside of Those Clusters
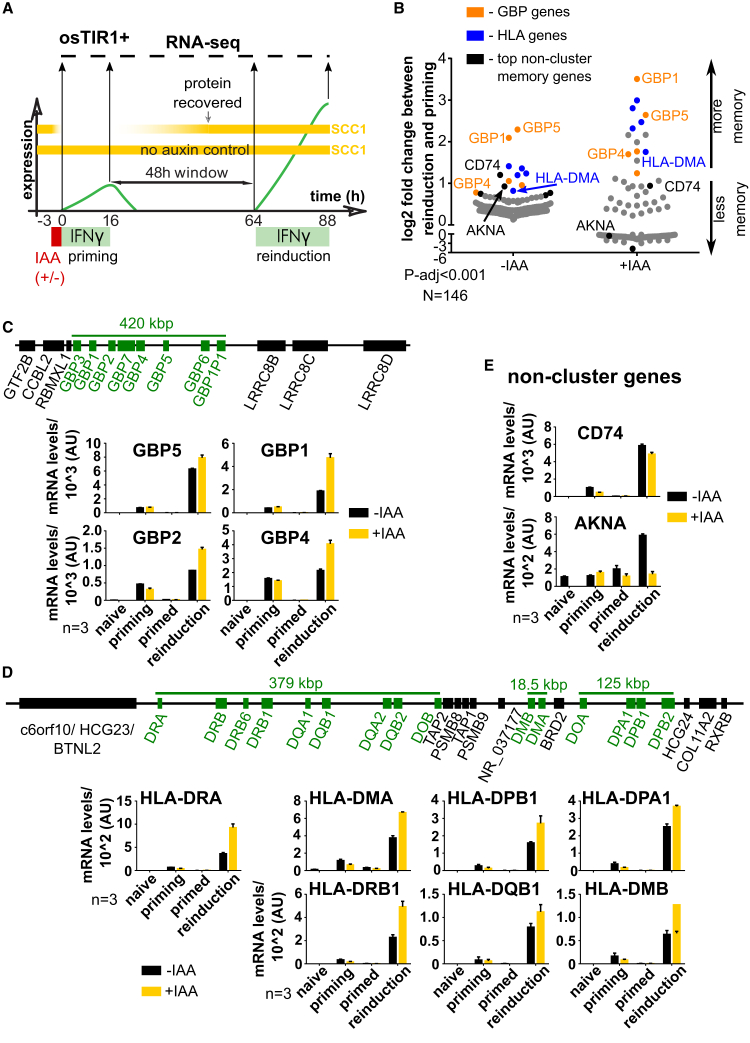
(A) Outline of a transcriptional memory experiment (analogous to Figure 1B) combined with auxin-mediated transient depletion of SCC1 during priming, analyzed by RNA-seq.
(B) HeLa Kyoto SCC1-EGFP-AID osTIR1-positive cells were subjected to the IFNγ and auxin treatment regimen outlined in (A) and processed for RNA-seq. The Log2 fold change for memory genes between reinduction (with or without auxin) and priming (without auxin) was plotted. “0” indicates no memory. The genes shown were selected based on a p-adj value below 0.001 as determined by the DESeq2 software (Love et al., 2014).
(C) Top: representation of the genomic structure of the GBP locus. Bottom: individual gene plots from the data described in (B). Error bars, SD; n = 3 biological replicates.
(D) Top: representation of the genomic structure of the HLA locus. Bottom: data presented as in (C) but for the HLA cluster genes.
(E) Data presented as in (C) but for the CD74 and AKNA genes.
See also Figures S4–S7.